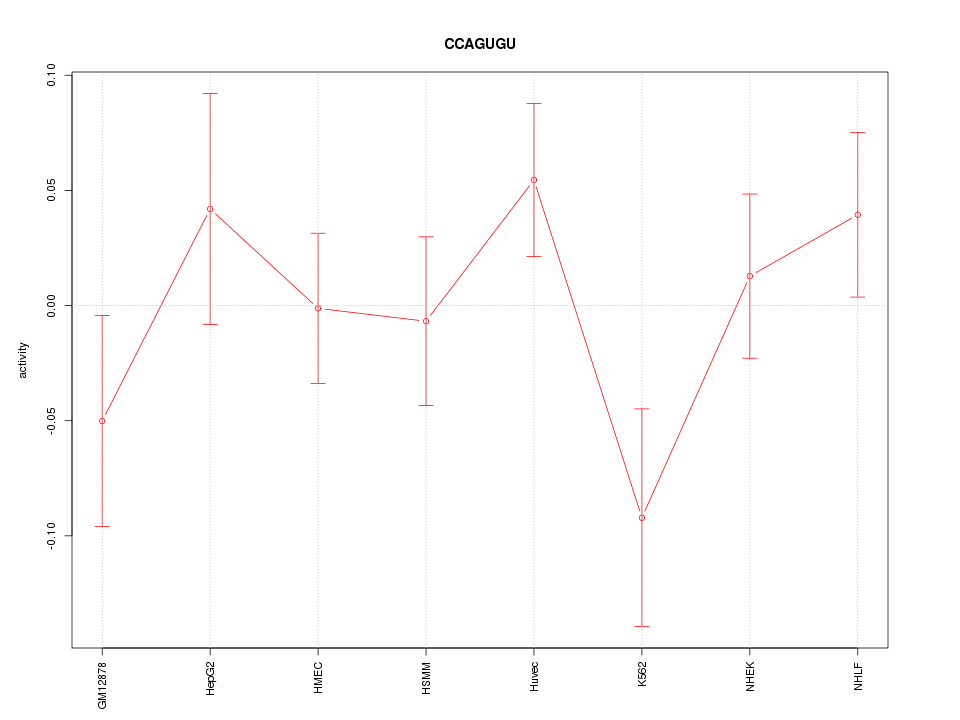
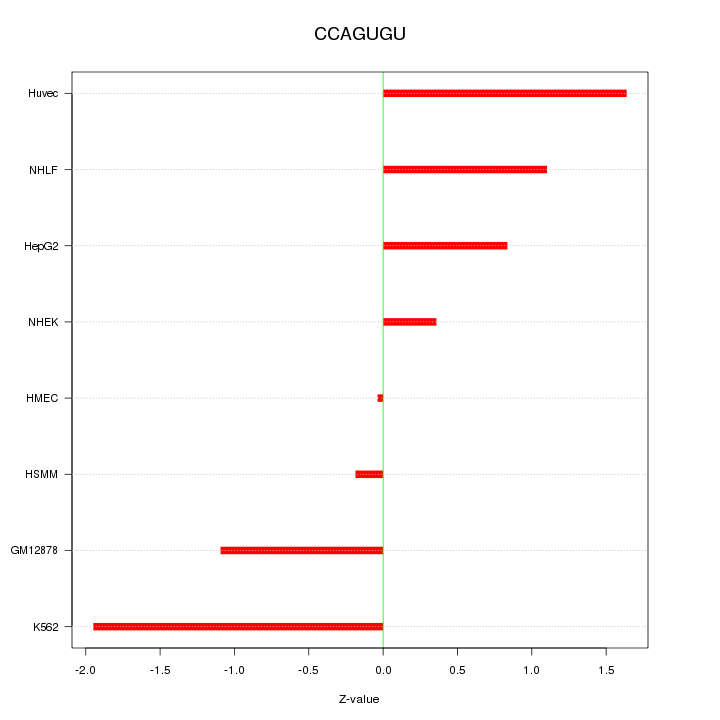
| 0.8 |
2.4 |
GO:0010757 |
chronological cell aging(GO:0001300) arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) negative regulation of plasminogen activation(GO:0010757) negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.5 |
1.6 |
GO:0061298 |
progesterone secretion(GO:0042701) retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.5 |
1.4 |
GO:0033278 |
cell proliferation in midbrain(GO:0033278) |
| 0.3 |
1.0 |
GO:0060370 |
susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.3 |
1.9 |
GO:0022617 |
extracellular matrix disassembly(GO:0022617) |
| 0.3 |
1.4 |
GO:0044854 |
membrane raft assembly(GO:0001765) nitric oxide homeostasis(GO:0033484) negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) membrane assembly(GO:0071709) |
| 0.3 |
1.0 |
GO:0033632 |
regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.2 |
0.7 |
GO:0046881 |
positive regulation of gonadotropin secretion(GO:0032278) positive regulation of follicle-stimulating hormone secretion(GO:0046881) Sertoli cell proliferation(GO:0060011) |
| 0.2 |
0.9 |
GO:0034959 |
neuropeptide catabolic process(GO:0010813) substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.2 |
0.4 |
GO:0035234 |
ectopic germ cell programmed cell death(GO:0035234) |
| 0.2 |
0.8 |
GO:0000212 |
meiotic spindle organization(GO:0000212) |
| 0.2 |
1.0 |
GO:0002328 |
pro-B cell differentiation(GO:0002328) |
| 0.2 |
0.7 |
GO:0050653 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.2 |
0.6 |
GO:1903332 |
regulation of protein folding(GO:1903332) |
| 0.1 |
0.3 |
GO:0045617 |
negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 |
0.6 |
GO:0010508 |
positive regulation of autophagy(GO:0010508) |
| 0.1 |
0.7 |
GO:0060037 |
pharyngeal system development(GO:0060037) |
| 0.1 |
0.4 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.1 |
0.3 |
GO:0060318 |
CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) definitive erythrocyte differentiation(GO:0060318) |
| 0.1 |
0.9 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 |
0.3 |
GO:0071109 |
superior temporal gyrus development(GO:0071109) |
| 0.1 |
0.7 |
GO:0040037 |
negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.1 |
0.7 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 |
0.2 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 |
0.6 |
GO:0048843 |
negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.1 |
0.6 |
GO:0007351 |
tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.1 |
0.2 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 |
0.3 |
GO:0032488 |
Cdc42 protein signal transduction(GO:0032488) |
| 0.1 |
0.4 |
GO:0000098 |
sulfur amino acid catabolic process(GO:0000098) |
| 0.1 |
0.4 |
GO:0051124 |
synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 |
0.2 |
GO:0061072 |
iris morphogenesis(GO:0061072) |
| 0.0 |
1.6 |
GO:0048009 |
insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 |
0.2 |
GO:0009439 |
cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.0 |
0.2 |
GO:0045722 |
response to muscle activity(GO:0014850) positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 |
0.4 |
GO:0060391 |
positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 |
0.5 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.0 |
0.2 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 |
0.3 |
GO:0051497 |
negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 |
0.2 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.0 |
0.2 |
GO:0032527 |
retrograde protein transport, ER to cytosol(GO:0030970) protein exit from endoplasmic reticulum(GO:0032527) |
| 0.0 |
0.9 |
GO:0030866 |
cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 |
0.1 |
GO:0090084 |
negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 |
0.4 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.0 |
0.2 |
GO:0050847 |
progesterone receptor signaling pathway(GO:0050847) |
| 0.0 |
0.1 |
GO:0006530 |
asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 |
0.3 |
GO:0032288 |
myelin assembly(GO:0032288) |
| 0.0 |
0.6 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.0 |
0.3 |
GO:0006596 |
polyamine biosynthetic process(GO:0006596) |
| 0.0 |
0.1 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) |
| 0.0 |
0.1 |
GO:0097352 |
autophagosome maturation(GO:0097352) |
| 0.0 |
0.0 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.0 |
0.1 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.1 |
GO:0014044 |
Schwann cell development(GO:0014044) myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 |
2.8 |
GO:0043123 |
positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 |
0.6 |
GO:0051101 |
regulation of DNA binding(GO:0051101) |
| 0.0 |
0.3 |
GO:0015949 |
nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 |
0.3 |
GO:0090630 |
activation of GTPase activity(GO:0090630) |
| 0.0 |
0.4 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.6 |
GO:0033692 |
cellular polysaccharide biosynthetic process(GO:0033692) |