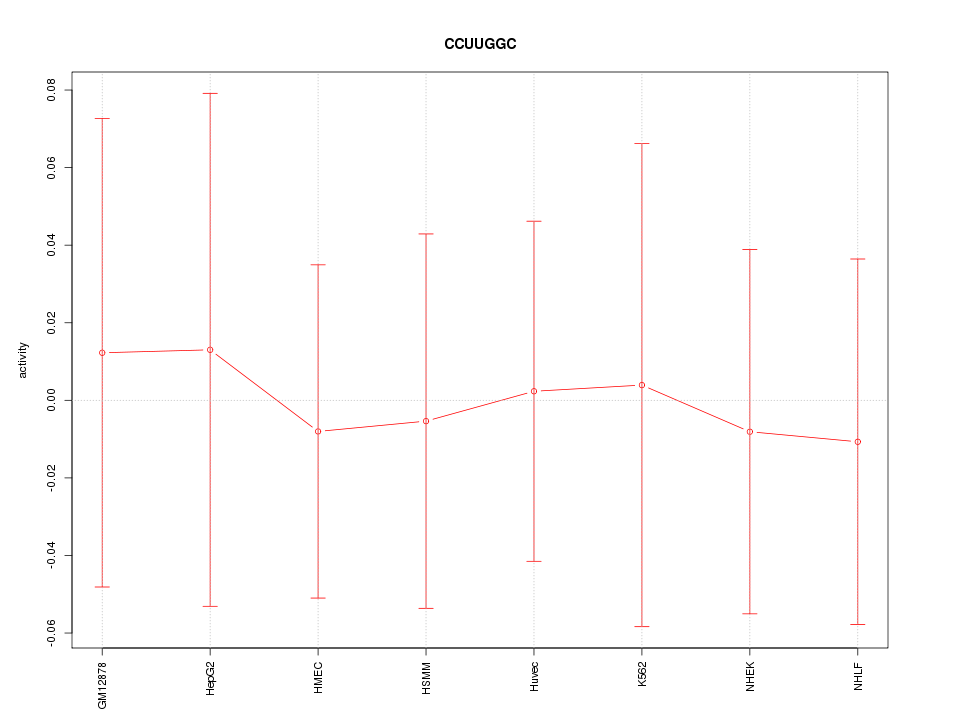
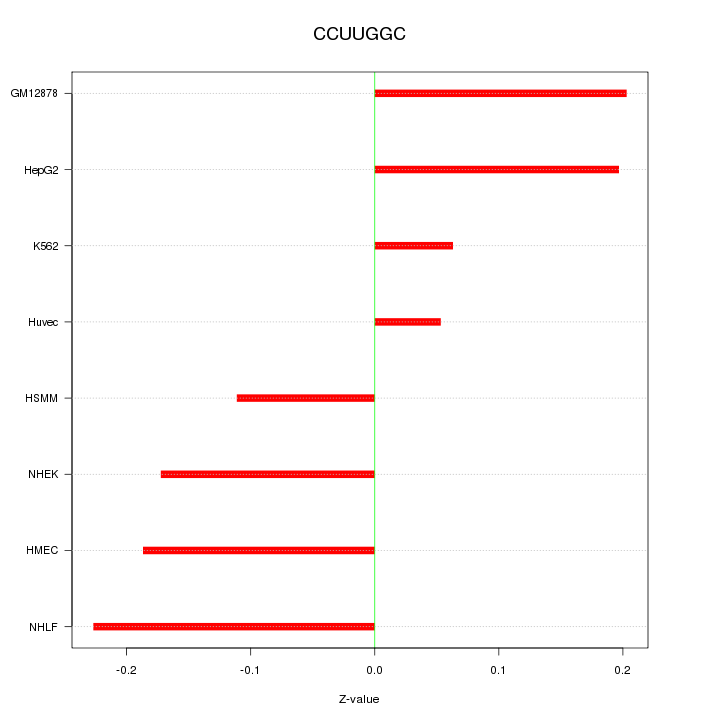
|
chr7_-_71801980
|
0.070
|
ENST00000329008.5
|
CALN1
|
calneuron 1
|
|
chr15_+_41952591
|
0.058
|
ENST00000566718.1
ENST00000219905.7
ENST00000389936.4
ENST00000545763.1
|
MGA
|
MGA, MAX dimerization protein
|
|
chr8_+_56014949
|
0.057
|
ENST00000327381.6
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4
|
|
chr20_+_37434329
|
0.051
|
ENST00000299824.1
ENST00000373331.2
|
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B
|
|
chr18_+_13218769
|
0.049
|
ENST00000399848.3
ENST00000361205.4
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4
|
|
chr19_+_926000
|
0.045
|
ENST00000263620.3
|
ARID3A
|
AT rich interactive domain 3A (BRIGHT-like)
|
|
chr8_-_8751068
|
0.044
|
ENST00000276282.6
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1
|
|
chr15_-_34628951
|
0.039
|
ENST00000397707.2
ENST00000560611.1
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6
|
|
chrY_+_15016725
|
0.037
|
ENST00000336079.3
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked
|
|
chr4_+_128554081
|
0.037
|
ENST00000335251.6
ENST00000296461.5
|
INTU
|
inturned planar cell polarity protein
|
|
chr16_-_89007491
|
0.036
|
ENST00000327483.5
ENST00000564416.1
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr1_+_214161272
|
0.035
|
ENST00000498508.2
ENST00000366958.4
|
PROX1
|
prospero homeobox 1
|
|
chr17_-_48785216
|
0.033
|
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40
|
|
chr2_-_27938593
|
0.032
|
ENST00000379677.2
|
AC074091.13
|
Uncharacterized protein
|
|
chr2_-_208634287
|
0.031
|
ENST00000295417.3
|
FZD5
|
frizzled family receptor 5
|
|
chr3_+_38495333
|
0.028
|
ENST00000352511.4
|
ACVR2B
|
activin A receptor, type IIB
|
|
chr3_-_53880401
|
0.028
|
ENST00000315251.6
|
CHDH
|
choline dehydrogenase
|
|
chrX_+_152953505
|
0.028
|
ENST00000253122.5
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter), member 8
|
|
chr11_+_75526212
|
0.027
|
ENST00000356136.3
|
UVRAG
|
UV radiation resistance associated
|
|
chr1_-_23886285
|
0.025
|
ENST00000374561.5
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein
|
|
chr19_+_42724423
|
0.025
|
ENST00000301215.3
ENST00000597945.1
|
ZNF526
|
zinc finger protein 526
|
|
chr1_+_11751748
|
0.025
|
ENST00000294485.5
|
DRAXIN
|
dorsal inhibitory axon guidance protein
|
|
chr2_+_61108650
|
0.022
|
ENST00000295025.8
|
REL
|
v-rel avian reticuloendotheliosis viral oncogene homolog
|
|
chr9_+_104161123
|
0.022
|
ENST00000374861.3
ENST00000339664.2
ENST00000259395.4
|
ZNF189
|
zinc finger protein 189
|
|
chr9_-_35665165
|
0.021
|
ENST00000343259.3
ENST00000378387.3
|
ARHGEF39
|
Rho guanine nucleotide exchange factor (GEF) 39
|
|
chr3_-_72496035
|
0.021
|
ENST00000477973.2
|
RYBP
|
RING1 and YY1 binding protein
|
|
chr2_-_96874553
|
0.020
|
ENST00000337288.5
ENST00000443962.1
|
STARD7
|
StAR-related lipid transfer (START) domain containing 7
|
|
chr8_-_82024290
|
0.020
|
ENST00000220597.4
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr17_-_8151353
|
0.020
|
ENST00000315684.8
|
CTC1
|
CTS telomere maintenance complex component 1
|
|
chr1_+_54519242
|
0.020
|
ENST00000234827.1
|
TCEANC2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2
|
|
chr22_+_50247449
|
0.019
|
ENST00000216268.5
|
ZBED4
|
zinc finger, BED-type containing 4
|
|
chr8_-_81083731
|
0.019
|
ENST00000379096.5
|
TPD52
|
tumor protein D52
|
|
chr10_+_48189612
|
0.018
|
ENST00000453919.1
|
AGAP9
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9
|
|
chr3_-_138553594
|
0.018
|
ENST00000477593.1
ENST00000483968.1
|
PIK3CB
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta
|
|
chr2_-_220094294
|
0.018
|
ENST00000436856.1
ENST00000428226.1
ENST00000409422.1
ENST00000431715.1
ENST00000457841.1
ENST00000439812.1
ENST00000361242.4
ENST00000396761.2
|
ATG9A
|
autophagy related 9A
|
|
chr10_+_72164135
|
0.018
|
ENST00000373218.4
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2
|
|
chr11_+_34127142
|
0.017
|
ENST00000257829.3
ENST00000531159.2
|
NAT10
|
N-acetyltransferase 10 (GCN5-related)
|
|
chr9_-_135996537
|
0.016
|
ENST00000372050.3
ENST00000372047.3
|
RALGDS
|
ral guanine nucleotide dissociation stimulator
|
|
chr17_-_76713100
|
0.016
|
ENST00000585509.1
|
CYTH1
|
cytohesin 1
|
|
chr17_-_60142609
|
0.015
|
ENST00000397786.2
|
MED13
|
mediator complex subunit 13
|
|
chr9_-_20622478
|
0.015
|
ENST00000355930.6
ENST00000380338.4
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3
|
|
chr14_-_95786200
|
0.015
|
ENST00000298912.4
|
CLMN
|
calmin (calponin-like, transmembrane)
|
|
chr6_-_99395787
|
0.015
|
ENST00000369244.2
ENST00000229971.1
|
FBXL4
|
F-box and leucine-rich repeat protein 4
|
|
chr1_-_89591749
|
0.014
|
ENST00000370466.3
|
GBP2
|
guanylate binding protein 2, interferon-inducible
|
|
chrX_-_50557014
|
0.014
|
ENST00000376020.2
|
SHROOM4
|
shroom family member 4
|
|
chr17_+_28804380
|
0.013
|
ENST00000225724.5
ENST00000451249.2
ENST00000467337.2
ENST00000581721.1
ENST00000414833.2
|
GOSR1
|
golgi SNAP receptor complex member 1
|
|
chr7_+_127292234
|
0.013
|
ENST00000354725.3
|
SND1
|
staphylococcal nuclease and tudor domain containing 1
|
|
chrX_+_77166172
|
0.013
|
ENST00000343533.5
ENST00000350425.4
ENST00000341514.6
|
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide
|
|
chr5_+_139781445
|
0.012
|
ENST00000394722.3
ENST00000532219.1
|
ANKHD1
ANKHD1-EIF4EBP3
|
ankyrin repeat and KH domain containing 1
ANKHD1-EIF4EBP3 readthrough
|
|
chr2_-_40679186
|
0.012
|
ENST00000406785.2
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr3_-_11762202
|
0.012
|
ENST00000445411.1
ENST00000404339.1
ENST00000273038.3
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr13_-_79177673
|
0.012
|
ENST00000377208.5
|
POU4F1
|
POU class 4 homeobox 1
|
|
chr16_-_15736953
|
0.012
|
ENST00000548025.1
ENST00000551742.1
ENST00000602337.1
ENST00000344181.3
ENST00000396368.3
|
KIAA0430
|
KIAA0430
|
|
chr1_+_201979645
|
0.012
|
ENST00000367284.5
ENST00000367283.3
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr2_-_85555385
|
0.011
|
ENST00000377386.3
|
TGOLN2
|
trans-golgi network protein 2
|
|
chr15_-_35261996
|
0.011
|
ENST00000156471.5
|
AQR
|
aquarius intron-binding spliceosomal factor
|
|
chr2_-_201936302
|
0.011
|
ENST00000453765.1
ENST00000452799.1
ENST00000446678.1
ENST00000418596.3
|
FAM126B
|
family with sequence similarity 126, member B
|
|
chr5_+_139927213
|
0.011
|
ENST00000310331.2
|
EIF4EBP3
|
eukaryotic translation initiation factor 4E binding protein 3
|
|
chr19_+_7953384
|
0.011
|
ENST00000306708.6
|
LRRC8E
|
leucine rich repeat containing 8 family, member E
|
|
chr5_-_37839782
|
0.010
|
ENST00000326524.2
ENST00000515058.1
|
GDNF
|
glial cell derived neurotrophic factor
|
|
chr10_+_111967345
|
0.009
|
ENST00000332674.5
ENST00000453116.1
|
MXI1
|
MAX interactor 1, dimerization protein
|
|
chr1_+_205012293
|
0.009
|
ENST00000331830.4
|
CNTN2
|
contactin 2 (axonal)
|
|
chr4_+_6784401
|
0.009
|
ENST00000425103.1
ENST00000307659.5
|
KIAA0232
|
KIAA0232
|
|
chr1_+_117602925
|
0.009
|
ENST00000369466.4
|
TTF2
|
transcription termination factor, RNA polymerase II
|
|
chr6_+_57182400
|
0.009
|
ENST00000607273.1
|
PRIM2
|
primase, DNA, polypeptide 2 (58kDa)
|
|
chr1_+_16330723
|
0.009
|
ENST00000329454.2
|
C1orf64
|
chromosome 1 open reading frame 64
|
|
chr8_+_76452097
|
0.008
|
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma
|
|
chr12_-_58240470
|
0.008
|
ENST00000548823.1
ENST00000398073.2
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2
|
|
chr16_-_71610985
|
0.008
|
ENST00000355962.4
|
TAT
|
tyrosine aminotransferase
|
|
chr17_-_45266542
|
0.008
|
ENST00000531206.1
ENST00000527547.1
ENST00000446365.2
ENST00000575483.1
ENST00000066544.3
|
CDC27
|
cell division cycle 27
|
|
chr3_+_183873098
|
0.008
|
ENST00000313143.3
|
DVL3
|
dishevelled segment polarity protein 3
|
|
chr17_-_62207485
|
0.007
|
ENST00000433197.3
|
ERN1
|
endoplasmic reticulum to nucleus signaling 1
|
|
chr15_+_44829255
|
0.007
|
ENST00000261868.5
ENST00000424492.3
|
EIF3J
|
eukaryotic translation initiation factor 3, subunit J
|
|
chr1_+_2985760
|
0.007
|
ENST00000378391.2
ENST00000514189.1
ENST00000270722.5
|
PRDM16
|
PR domain containing 16
|
|
chrX_+_41192595
|
0.007
|
ENST00000399959.2
|
DDX3X
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked
|
|
chr16_+_81348528
|
0.007
|
ENST00000568107.2
|
GAN
|
gigaxonin
|
|
chr11_-_82782861
|
0.007
|
ENST00000524635.1
ENST00000526205.1
ENST00000527633.1
ENST00000533486.1
ENST00000533276.2
|
RAB30
|
RAB30, member RAS oncogene family
|
|
chr16_-_18937726
|
0.007
|
ENST00000389467.3
ENST00000446231.2
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase
|
|
chr1_+_25757376
|
0.006
|
ENST00000399766.3
ENST00000399763.3
ENST00000374343.4
|
TMEM57
|
transmembrane protein 57
|
|
chrX_+_122993827
|
0.006
|
ENST00000371199.3
|
XIAP
|
X-linked inhibitor of apoptosis
|
|
chr3_-_33686743
|
0.005
|
ENST00000333778.6
ENST00000539981.1
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr11_+_65819802
|
0.005
|
ENST00000528302.1
ENST00000322535.6
ENST00000524627.1
ENST00000533595.1
ENST00000530322.1
|
SF3B2
|
splicing factor 3b, subunit 2, 145kDa
|
|
chr16_+_50775948
|
0.005
|
ENST00000569681.1
ENST00000569418.1
ENST00000540145.1
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|
|
chr8_+_128426535
|
0.005
|
ENST00000465342.2
|
POU5F1B
|
POU class 5 homeobox 1B
|
|
chr17_+_8213590
|
0.004
|
ENST00000361926.3
|
ARHGEF15
|
Rho guanine nucleotide exchange factor (GEF) 15
|
|
chr6_+_50786414
|
0.004
|
ENST00000344788.3
ENST00000393655.3
ENST00000263046.4
|
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta)
|
|
chr7_-_92463210
|
0.004
|
ENST00000265734.4
|
CDK6
|
cyclin-dependent kinase 6
|
|
chr12_+_70636765
|
0.004
|
ENST00000552231.1
ENST00000229195.3
ENST00000547780.1
ENST00000418359.3
|
CNOT2
|
CCR4-NOT transcription complex, subunit 2
|
|
chr2_+_159313452
|
0.004
|
ENST00000389757.3
ENST00000389759.3
|
PKP4
|
plakophilin 4
|
|
chr3_-_114790179
|
0.003
|
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr9_-_127269661
|
0.003
|
ENST00000373588.4
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1
|
|
chr2_+_168725458
|
0.003
|
ENST00000392690.3
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1
|
|
chr15_+_75118888
|
0.002
|
ENST00000395018.4
|
CPLX3
|
complexin 3
|
|
chr11_+_61447845
|
0.002
|
ENST00000257215.5
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chr20_+_33814457
|
0.002
|
ENST00000246186.6
|
MMP24
|
matrix metallopeptidase 24 (membrane-inserted)
|
|
chr2_+_219264466
|
0.002
|
ENST00000273062.2
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1
|
|
chr17_+_47865917
|
0.002
|
ENST00000259021.4
ENST00000454930.2
ENST00000509773.1
ENST00000510819.1
ENST00000424009.2
|
KAT7
|
K(lysine) acetyltransferase 7
|
|
chr16_-_46865047
|
0.001
|
ENST00000394806.2
|
C16orf87
|
chromosome 16 open reading frame 87
|
|
chr1_+_178995021
|
0.001
|
ENST00000263733.4
|
FAM20B
|
family with sequence similarity 20, member B
|
|
chr6_-_91006461
|
0.001
|
ENST00000257749.4
ENST00000343122.3
ENST00000406998.2
ENST00000453877.1
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr17_-_73179046
|
0.000
|
ENST00000314523.7
ENST00000420826.2
|
SUMO2
|
small ubiquitin-like modifier 2
|
|
chr11_+_120207787
|
0.000
|
ENST00000397843.2
ENST00000356641.3
|
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12
|
|
chr8_+_37594130
|
0.000
|
ENST00000518526.1
ENST00000523887.1
ENST00000276461.5
|
ERLIN2
|
ER lipid raft associated 2
|