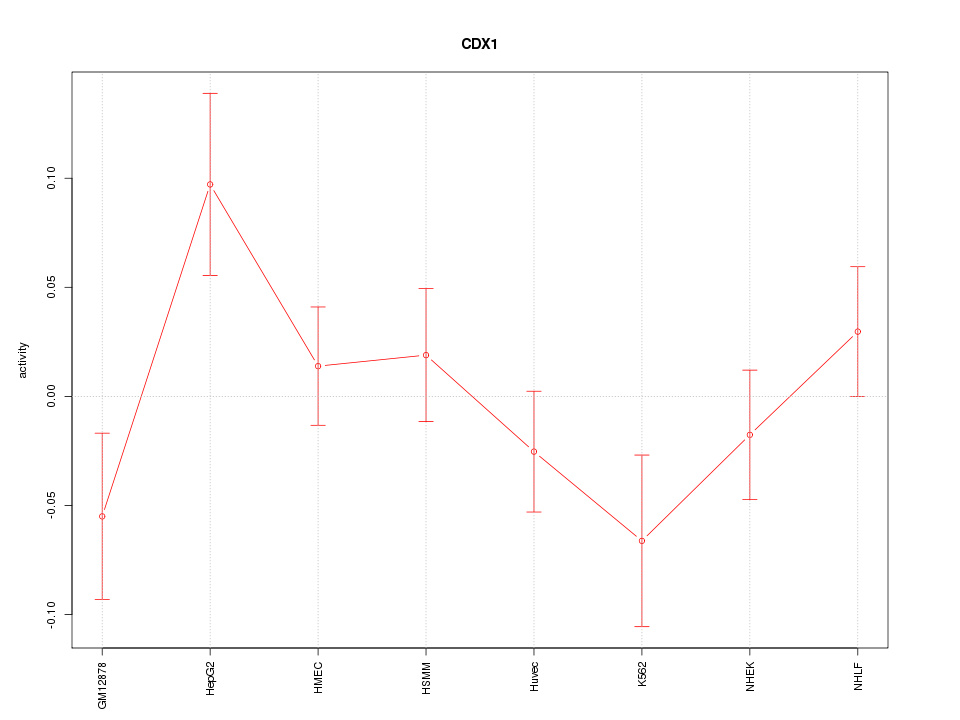
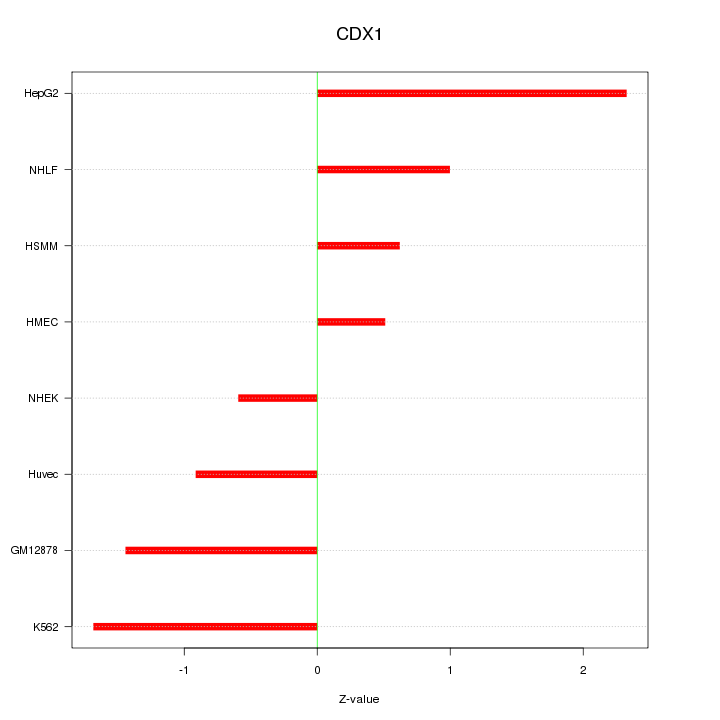
Motif ID: CDX1
Z-value: 1.283
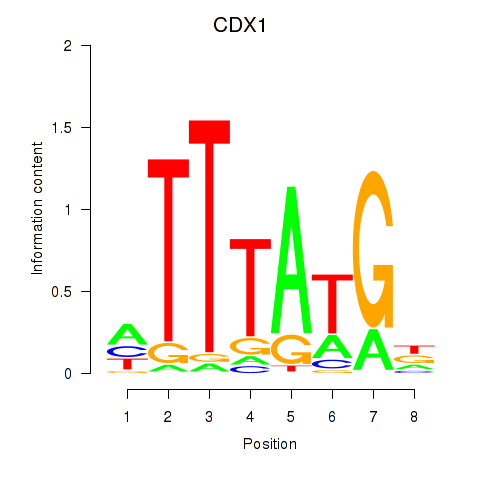
Transcription factors associated with CDX1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| CDX1 | ENSG00000113722.12 | CDX1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.7 | 2.1 | GO:0061117 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.7 | 2.0 | GO:0001907 | cytolysis by symbiont of host cells(GO:0001897) killing by symbiont of host cells(GO:0001907) hemolysis by symbiont of host erythrocytes(GO:0019836) disruption by symbiont of host cell(GO:0044004) hemolysis in other organism(GO:0044179) positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) cytolysis in other organism(GO:0051715) cytolysis in other organism involved in symbiotic interaction(GO:0051801) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.7 | 3.9 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.5 | 2.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.4 | 4.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.3 | 2.1 | GO:0060414 | aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 2.0 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.2 | 0.5 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.2 | 1.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.2 | 1.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.6 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.1 | 0.7 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.4 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 1.6 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 6.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.8 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 | 0.5 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.1 | 0.4 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) long-term synaptic potentiation(GO:0060291) |
| 0.1 | 0.3 | GO:0043371 | negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.1 | 2.1 | GO:0042094 | interleukin-2 biosynthetic process(GO:0042094) |
| 0.1 | 1.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.1 | 3.3 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 0.2 | GO:0071224 | cell surface pattern recognition receptor signaling pathway(GO:0002752) interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) cellular response to peptidoglycan(GO:0071224) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.1 | 0.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 1.2 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.6 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.3 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.3 | GO:0071220 | response to bacterial lipoprotein(GO:0032493) response to bacterial lipopeptide(GO:0070339) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) |
| 0.0 | 0.1 | GO:0031034 | skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0071801 | podosome assembly(GO:0071800) regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.7 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.1 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.6 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.3 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.3 | GO:0051205 | respiratory chain complex IV assembly(GO:0008535) protein insertion into membrane(GO:0051205) |
| 0.0 | 3.4 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.5 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.9 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.4 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.2 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 1.6 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 1.1 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 1.0 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.0 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.7 | 4.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 2.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.5 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.1 | 1.7 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 1.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 6.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 0.2 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.3 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.9 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 3.4 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.0 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.5 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 1.0 | 4.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.5 | 2.2 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.5 | 1.6 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.4 | 2.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.4 | 1.1 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.4 | 5.0 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.3 | 0.8 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.2 | 2.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 1.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 6.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 2.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 2.1 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 0.4 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.7 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 3.8 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.1 | 0.5 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.1 | 0.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.3 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.1 | 1.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) lipopeptide binding(GO:0071723) |
| 0.1 | 0.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.6 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 1.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.1 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 3.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 2.2 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.7 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.7 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |