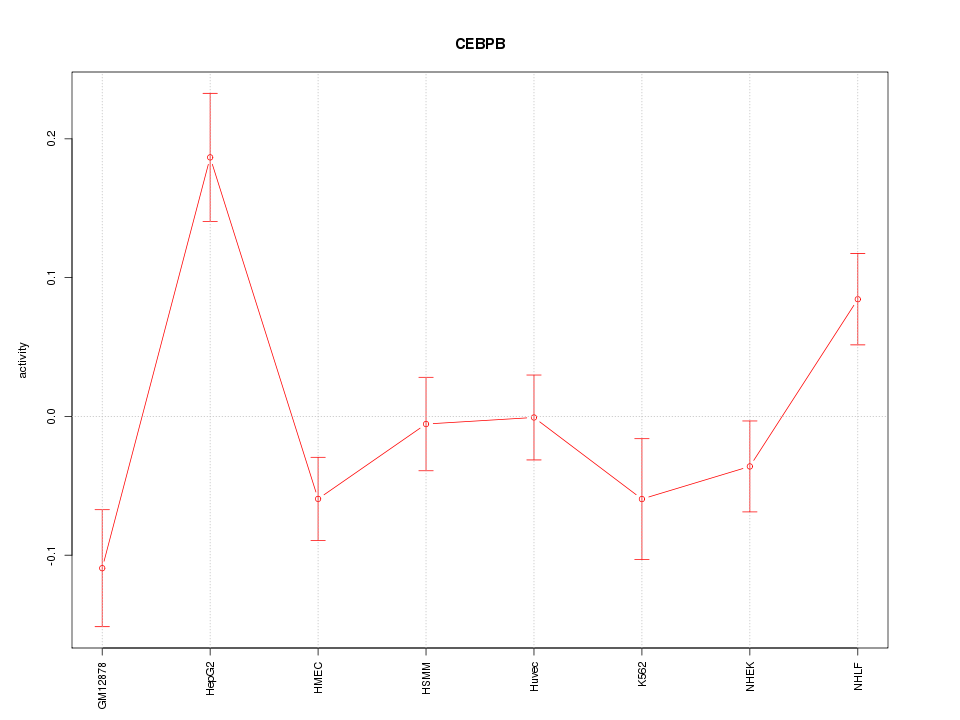
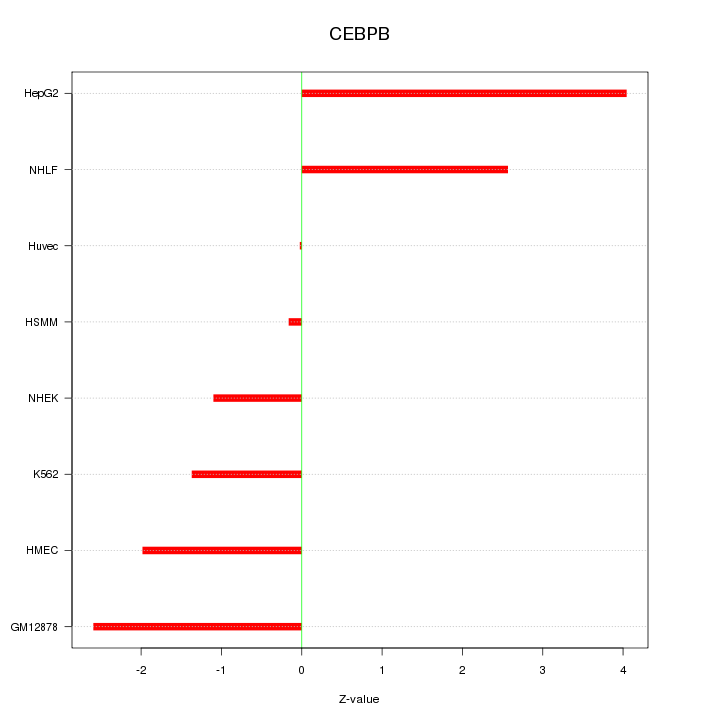
Motif ID: CEBPB
Z-value: 2.143
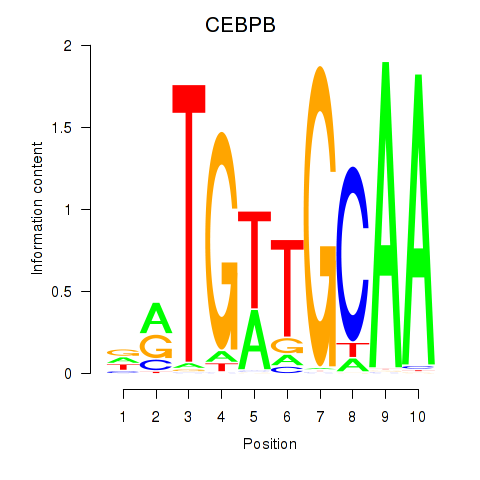
Transcription factors associated with CEBPB:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| CEBPB | ENSG00000172216.4 | CEBPB |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.3 | GO:0007614 | short-term memory(GO:0007614) |
| 1.2 | 6.2 | GO:1901031 | regulation of cellular response to oxidative stress(GO:1900407) regulation of response to reactive oxygen species(GO:1901031) regulation of response to oxidative stress(GO:1902882) |
| 1.2 | 3.5 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 1.0 | 7.7 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.9 | 2.8 | GO:0043418 | cysteine biosynthetic process from serine(GO:0006535) homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) transsulfuration(GO:0019346) homocysteine catabolic process(GO:0043418) |
| 0.9 | 2.8 | GO:0006788 | heme oxidation(GO:0006788) smooth muscle hyperplasia(GO:0014806) muscle hyperplasia(GO:0014900) |
| 0.8 | 2.3 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.6 | 3.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) regulation of cholesterol transporter activity(GO:0060694) |
| 0.5 | 1.9 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.5 | 1.4 | GO:0070672 | response to interleukin-12(GO:0070671) response to interleukin-15(GO:0070672) |
| 0.4 | 2.8 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.4 | 1.1 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.3 | 6.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 1.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.3 | 4.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.3 | 0.8 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.3 | 1.6 | GO:0008228 | opsonization(GO:0008228) |
| 0.3 | 0.8 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.3 | 3.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.2 | 1.5 | GO:0015811 | sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.2 | 3.3 | GO:0046697 | decidualization(GO:0046697) |
| 0.2 | 1.4 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 0.5 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.4 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.2 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.5 | GO:0048550 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) negative regulation of pinocytosis(GO:0048550) |
| 0.1 | 0.2 | GO:0001554 | luteolysis(GO:0001554) |
| 0.1 | 2.6 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 1.7 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 0.2 | GO:0003093 | regulation of glomerular filtration(GO:0003093) glomerular filtration(GO:0003094) renal filtration(GO:0097205) regulation of renal system process(GO:0098801) |
| 0.1 | 3.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.3 | GO:0017014 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.1 | 1.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.5 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 0.6 | GO:0030147 | obsolete natriuresis(GO:0030147) |
| 0.1 | 0.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.7 | GO:0042977 | activation of JAK2 kinase activity(GO:0042977) |
| 0.0 | 0.9 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 0.8 | GO:0001960 | negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.0 | 4.4 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.0 | 1.0 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 3.0 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.0 | 0.4 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 0.1 | GO:0070942 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) |
| 0.0 | 1.6 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.1 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.5 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.6 | GO:0043525 | positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.0 | 0.3 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 1.2 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 0.5 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 2.7 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.0 | 0.2 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.2 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.1 | GO:0002675 | positive regulation of acute inflammatory response(GO:0002675) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.9 | 9.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.7 | 2.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 7.7 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 1.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 4.7 | GO:0042645 | mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 11.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 2.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 2.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.9 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 1.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.4 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 2.8 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 2.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 1.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 1.9 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 6.8 | GO:0005625 | obsolete soluble fraction(GO:0005625) |
| 0.0 | 1.5 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 22.9 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.1 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.7 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.9 | 2.8 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.9 | 2.8 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.9 | 2.7 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.7 | 6.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.6 | 2.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.4 | 2.8 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.4 | 2.7 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 2.4 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.3 | 3.0 | GO:1901567 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.3 | 1.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.3 | 0.8 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.3 | 0.8 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.2 | 3.8 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.2 | 5.0 | GO:0043499 | obsolete eukaryotic cell surface binding(GO:0043499) |
| 0.2 | 0.7 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.2 | 2.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 1.7 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 2.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.1 | 0.5 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 0.6 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 1.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.2 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.1 | 0.3 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 10.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.0 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 4.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 1.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 0.3 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 1.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 6.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 6.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 3.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.5 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.0 | 2.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 1.7 | GO:0004221 | obsolete ubiquitin thiolesterase activity(GO:0004221) |
| 0.0 | 5.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.4 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 1.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.7 | GO:0016684 | peroxidase activity(GO:0004601) oxidoreductase activity, acting on peroxide as acceptor(GO:0016684) |
| 0.0 | 1.4 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.4 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 1.6 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 3.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.2 | GO:0005109 | frizzled binding(GO:0005109) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.1 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.4 | ST_G_ALPHA_I_PATHWAY | G alpha i Pathway |
| 0.0 | 2.0 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.3 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |