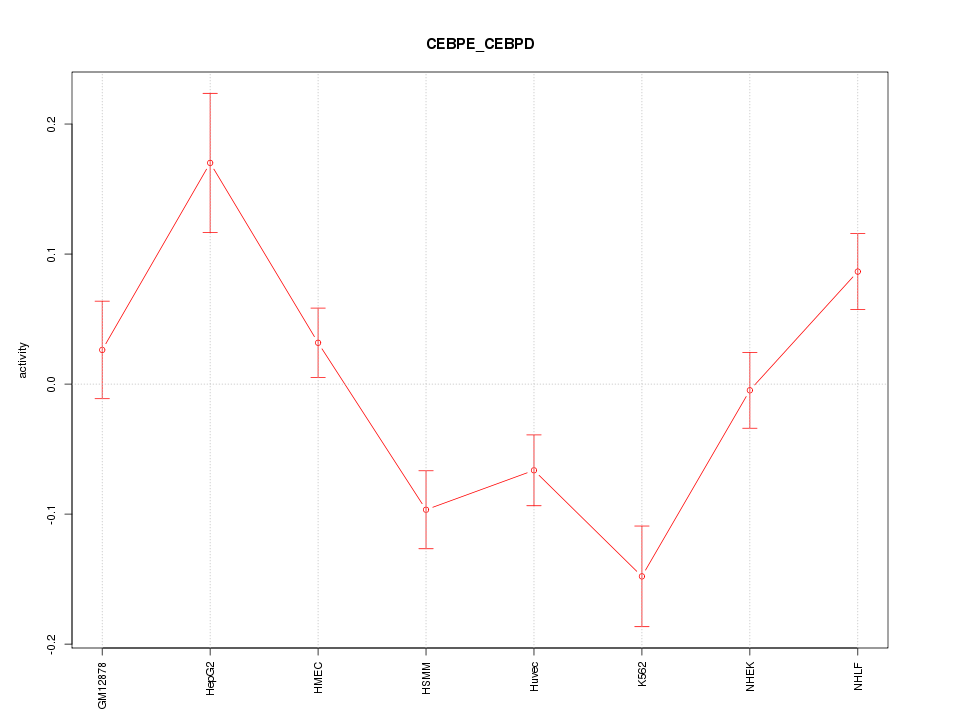
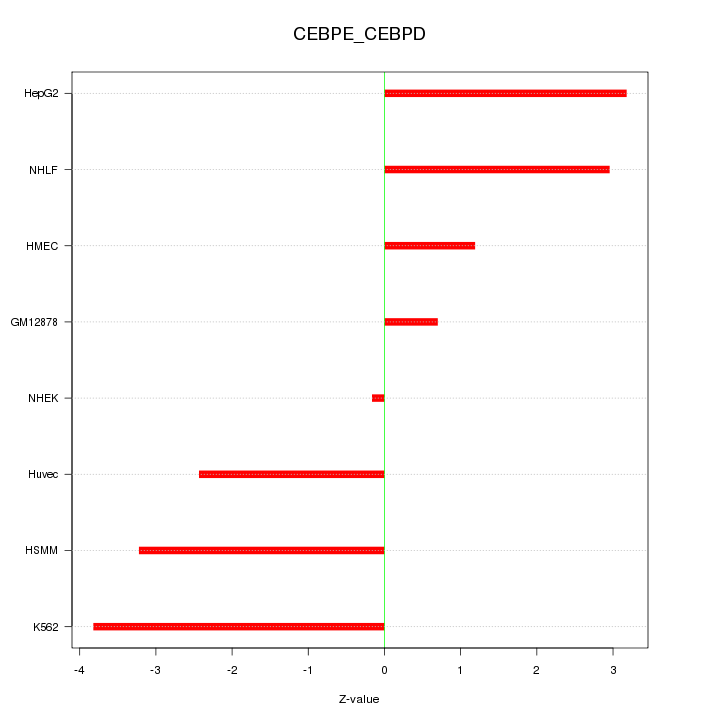
Motif ID: CEBPE_CEBPD
Z-value: 2.542
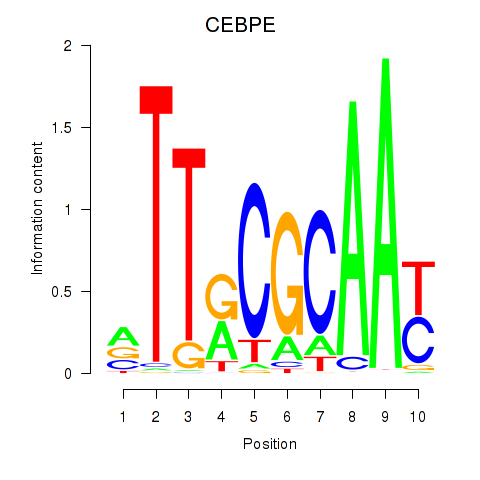
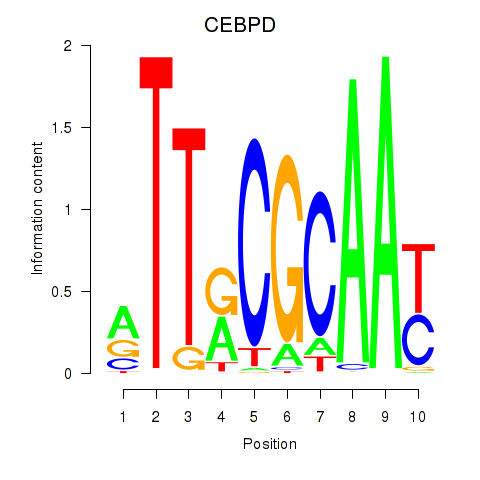
Transcription factors associated with CEBPE_CEBPD:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| CEBPD | ENSG00000221869.4 | CEBPD |
| CEBPE | ENSG00000092067.5 | CEBPE |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 18.0 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 1.9 | 7.5 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) positive regulation of activation of membrane attack complex(GO:0001970) positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 1.5 | 4.5 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 1.1 | 3.3 | GO:0002384 | hepatic immune response(GO:0002384) |
| 1.1 | 7.6 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 1.0 | 3.9 | GO:0001705 | ectoderm formation(GO:0001705) central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.9 | 3.6 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.9 | 4.3 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) regulation of cholesterol transporter activity(GO:0060694) |
| 0.9 | 3.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.8 | 5.3 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.5 | 5.4 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.5 | 3.6 | GO:0034695 | response to prostaglandin E(GO:0034695) |
| 0.4 | 1.2 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.4 | 2.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.3 | 6.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.3 | 6.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 0.9 | GO:0006740 | glyoxylate cycle(GO:0006097) NADPH regeneration(GO:0006740) |
| 0.3 | 2.1 | GO:0048296 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.3 | 3.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 9.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.2 | 1.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 1.1 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.2 | 10.9 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.2 | 3.0 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.2 | 1.1 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.2 | 10.0 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.2 | 1.6 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.2 | 1.7 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.2 | 2.0 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.2 | 0.5 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.1 | 1.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.4 | GO:0000114 | obsolete regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0000114) |
| 0.1 | 0.4 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 1.4 | GO:0030147 | obsolete natriuresis(GO:0030147) |
| 0.1 | 0.3 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.5 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.1 | 0.3 | GO:0061117 | cardiac left ventricle formation(GO:0003218) negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.1 | 0.5 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 | 6.1 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.1 | 1.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.4 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.1 | 2.8 | GO:0033572 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 | 5.0 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.1 | 1.4 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 0.3 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 0.5 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 5.3 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.0 | 1.0 | GO:0009070 | serine family amino acid biosynthetic process(GO:0009070) |
| 0.0 | 0.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 1.0 | GO:0001960 | negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.0 | 1.9 | GO:0007045 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 | 0.0 | GO:0071639 | heterotypic cell-cell adhesion(GO:0034113) regulation of heterotypic cell-cell adhesion(GO:0034114) monocyte chemotactic protein-1 production(GO:0071605) regulation of monocyte chemotactic protein-1 production(GO:0071637) positive regulation of monocyte chemotactic protein-1 production(GO:0071639) |
| 0.0 | 2.5 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 1.7 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.3 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.6 | GO:0045069 | regulation of viral genome replication(GO:0045069) |
| 0.0 | 0.5 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) cellular response to unfolded protein(GO:0034620) |
| 0.0 | 0.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 2.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.5 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.4 | GO:0042994 | cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 | 0.4 | GO:0031055 | DNA replication-independent nucleosome assembly(GO:0006336) chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) DNA replication-independent nucleosome organization(GO:0034724) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 1.4 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.0 | 0.3 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.2 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 2.6 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.6 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.2 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 0.1 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0071223 | response to lipoteichoic acid(GO:0070391) cellular response to lipoteichoic acid(GO:0071223) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 21.5 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.8 | 3.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.4 | 4.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 5.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 3.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 2.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 5.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 4.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 43.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.6 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 3.7 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.3 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 4.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 7.0 | GO:0005625 | obsolete soluble fraction(GO:0005625) |
| 0.0 | 0.1 | GO:0001673 | male germ cell nucleus(GO:0001673) germ cell nucleus(GO:0043073) |
| 0.0 | 0.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 4.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 9.0 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.0 | 3.8 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.9 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.6 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 1.4 | 6.9 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.3 | 5.3 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 1.1 | 18.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 1.1 | 4.5 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 1.1 | 3.3 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.8 | 2.5 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.8 | 7.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.7 | 6.4 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.7 | 2.0 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.6 | 1.9 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.5 | 3.6 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.5 | 4.3 | GO:1901567 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.4 | 5.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 1.0 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.3 | 1.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.3 | 0.9 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.3 | 1.7 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.3 | 6.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 1.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.3 | 2.5 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 1.2 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.2 | 3.9 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.2 | 0.7 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.2 | 1.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 1.6 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 0.3 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 5.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.8 | GO:0043499 | obsolete eukaryotic cell surface binding(GO:0043499) |
| 0.1 | 0.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.5 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 2.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 12.4 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 1.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.9 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 3.4 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.1 | GO:0070891 | peptidoglycan receptor activity(GO:0016019) lipoteichoic acid binding(GO:0070891) |
| 0.0 | 1.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.5 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 6.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 1.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.6 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.3 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 5.2 | GO:0030528 | obsolete transcription regulator activity(GO:0030528) |
| 0.0 | 0.7 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 1.4 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 1.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | ST_STAT3_PATHWAY | STAT3 Pathway |
| 0.1 | 6.3 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.1 | 2.8 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.4 | ST_GAQ_PATHWAY | G alpha q Pathway |