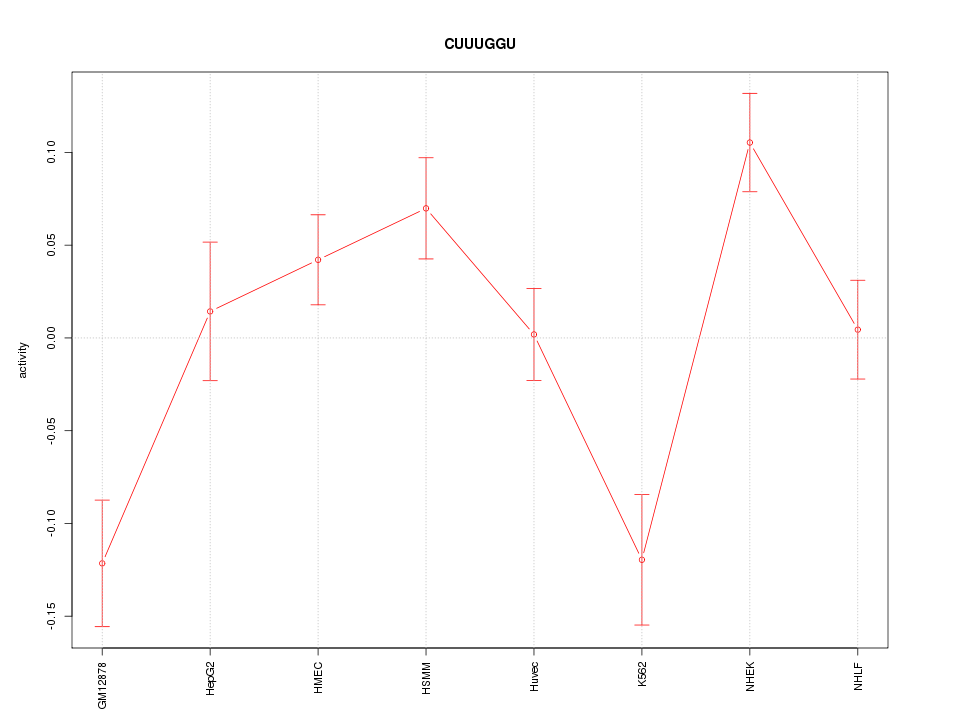
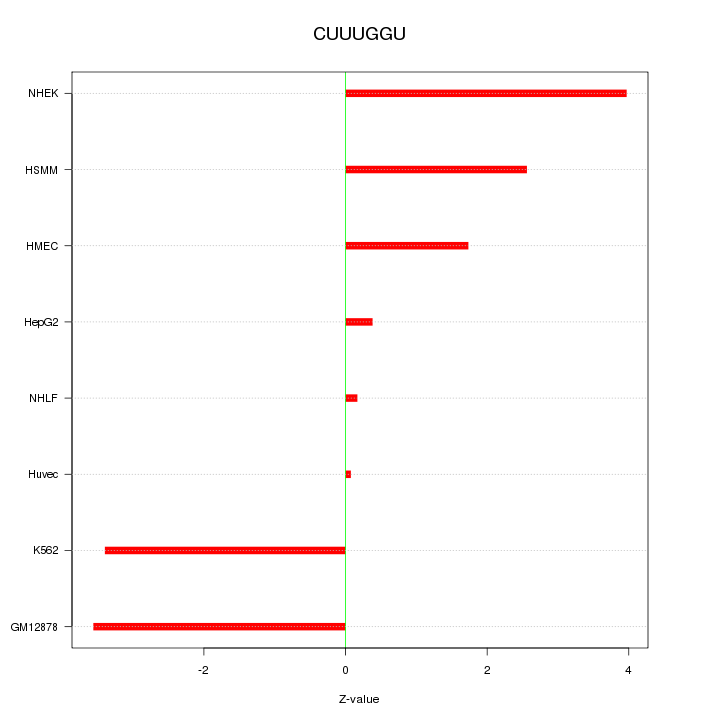
Motif ID: CUUUGGU
Z-value: 2.494

Mature miRNA associated with seed CUUUGGU:
| Name | miRBase Accession |
|---|---|
| hsa-miR-9-5p | MIMAT0000441 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.9 | 2.7 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.9 | 2.6 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) dermatan sulfate biosynthetic process(GO:0030208) |
| 0.8 | 2.4 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.7 | 1.4 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.6 | 1.9 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.6 | 1.8 | GO:0043276 | anoikis(GO:0043276) |
| 0.5 | 1.5 | GO:0048627 | myoblast development(GO:0048627) |
| 0.5 | 2.5 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.5 | 1.4 | GO:0003176 | aortic valve development(GO:0003176) aortic valve morphogenesis(GO:0003180) notochord formation(GO:0014028) |
| 0.4 | 3.8 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.4 | 1.5 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.4 | 3.3 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.4 | 2.5 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.3 | 4.5 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.3 | 2.3 | GO:0060414 | aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 1.9 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.3 | 0.9 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.3 | 2.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.3 | 3.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.3 | 0.9 | GO:0002124 | territorial aggressive behavior(GO:0002124) brainstem development(GO:0003360) vocalization behavior(GO:0071625) |
| 0.3 | 0.9 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.3 | 1.8 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.3 | 3.3 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.3 | 5.1 | GO:0001960 | negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.2 | 0.2 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.2 | 3.6 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.2 | 0.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.2 | 0.9 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.2 | 3.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.2 | 0.6 | GO:0046520 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.2 | 1.9 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 0.6 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.2 | 0.8 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 1.8 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.2 | 1.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.2 | 0.4 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.2 | 0.9 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.2 | 8.9 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.2 | 0.5 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.2 | 1.5 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.2 | 0.9 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.2 | 2.0 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.2 | 0.6 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.4 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.1 | 0.4 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 2.8 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 2.2 | GO:0002011 | morphogenesis of an epithelial sheet(GO:0002011) |
| 0.1 | 0.7 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 0.2 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 | 1.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 0.4 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.1 | 2.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.5 | GO:0034204 | Cdc42 protein signal transduction(GO:0032488) lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 1.6 | GO:0060438 | trachea development(GO:0060438) |
| 0.1 | 0.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.5 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 2.1 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 2.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 0.3 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.3 | GO:0001999 | renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) |
| 0.1 | 0.5 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 0.5 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.5 | GO:0010833 | telomere maintenance via telomere lengthening(GO:0010833) |
| 0.1 | 2.2 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.1 | 0.8 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.1 | 0.4 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 7.0 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.4 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.5 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 0.2 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 0.3 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.1 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 1.3 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.8 | GO:0018298 | detection of visible light(GO:0009584) protein-chromophore linkage(GO:0018298) |
| 0.0 | 4.5 | GO:0071772 | BMP signaling pathway(GO:0030509) response to BMP(GO:0071772) cellular response to BMP stimulus(GO:0071773) |
| 0.0 | 0.2 | GO:0097479 | synaptic vesicle transport(GO:0048489) synaptic vesicle localization(GO:0097479) establishment of synaptic vesicle localization(GO:0097480) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 2.4 | GO:0001843 | neural tube closure(GO:0001843) primary neural tube formation(GO:0014020) tube closure(GO:0060606) |
| 0.0 | 0.4 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 1.6 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.3 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 1.7 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 2.0 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.3 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.4 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.8 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.5 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.0 | 0.5 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 2.2 | GO:0034332 | adherens junction organization(GO:0034332) |
| 0.0 | 0.1 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.6 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.2 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 1.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.0 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.7 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 2.7 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 2.6 | GO:0019319 | hexose biosynthetic process(GO:0019319) |
| 0.0 | 0.6 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 1.3 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) negative regulation of cell projection organization(GO:0031345) |
| 0.0 | 0.2 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.3 | GO:0017156 | calcium ion regulated exocytosis(GO:0017156) |
| 0.0 | 1.6 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 0.1 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.2 | GO:0032844 | regulation of homeostatic process(GO:0032844) |
| 0.0 | 0.6 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.4 | GO:1903305 | regulation of calcium ion-dependent exocytosis(GO:0017158) regulation of regulated secretory pathway(GO:1903305) |
| 0.0 | 0.3 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.1 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.0 | 0.0 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 1.1 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.3 | GO:0019400 | glycerol metabolic process(GO:0006071) alditol metabolic process(GO:0019400) |
| 0.0 | 0.0 | GO:0021604 | rhombomere 4 development(GO:0021570) cranial nerve structural organization(GO:0021604) facial nerve morphogenesis(GO:0021610) facial nerve structural organization(GO:0021612) facial nucleus development(GO:0021754) |
| 0.0 | 0.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.6 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.3 | GO:0001932 | regulation of protein phosphorylation(GO:0001932) |
| 0.0 | 0.1 | GO:0042977 | activation of JAK2 kinase activity(GO:0042977) |
| 0.0 | 0.2 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) |
| 0.0 | 0.0 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.0 | 0.5 | GO:0042990 | regulation of transcription factor import into nucleus(GO:0042990) transcription factor import into nucleus(GO:0042991) |
| 0.0 | 0.6 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.6 | 1.7 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.5 | 3.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.4 | 1.8 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.4 | 2.0 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.4 | 2.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.3 | 2.6 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.2 | 3.8 | GO:0001527 | microfibril(GO:0001527) |
| 0.2 | 2.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 1.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 1.5 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.2 | 1.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 1.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.5 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.5 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.1 | 0.4 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 5.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 0.9 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.9 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.9 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 10.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.4 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 4.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 16.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 2.8 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 0.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.8 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.3 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 1.1 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 1.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 1.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 2.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.5 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 2.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 3.2 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 2.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 2.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 3.3 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 1.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 2.3 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 1.2 | 3.7 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.9 | 4.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.8 | 3.4 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) protein phosphatase activator activity(GO:0072542) |
| 0.8 | 3.2 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.6 | 3.8 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.6 | 3.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.6 | 1.7 | GO:0048184 | obsolete follistatin binding(GO:0048184) |
| 0.5 | 2.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.5 | 2.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.5 | 2.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.4 | 1.8 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.4 | 1.7 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.4 | 1.2 | GO:0050816 | beta-2 adrenergic receptor binding(GO:0031698) phosphothreonine binding(GO:0050816) |
| 0.3 | 3.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.3 | 0.9 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.3 | 2.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.3 | 5.9 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.3 | 1.3 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.3 | 1.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.3 | 2.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 2.6 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.2 | 1.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 0.6 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.2 | 2.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 0.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 1.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.2 | 4.6 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 1.0 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 0.5 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 1.4 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.2 | 1.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 0.9 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.1 | 0.4 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.1 | 5.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 2.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.5 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 2.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.5 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.1 | 1.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 1.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 2.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.2 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.1 | 0.4 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 2.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.5 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 1.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.5 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 1.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.4 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.3 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 1.0 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.1 | 7.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 4.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.9 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 1.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 7.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 2.9 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 1.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.5 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.3 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.0 | 0.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 2.3 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 4.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.4 | GO:0004437 | obsolete inositol or phosphatidylinositol phosphatase activity(GO:0004437) |
| 0.0 | 0.5 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 3.1 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 2.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.2 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 3.0 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.2 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.1 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.5 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.8 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.8 | SA_TRKA_RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 2.5 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.2 | ST_JAK_STAT_PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.7 | ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | PI3K Pathway |