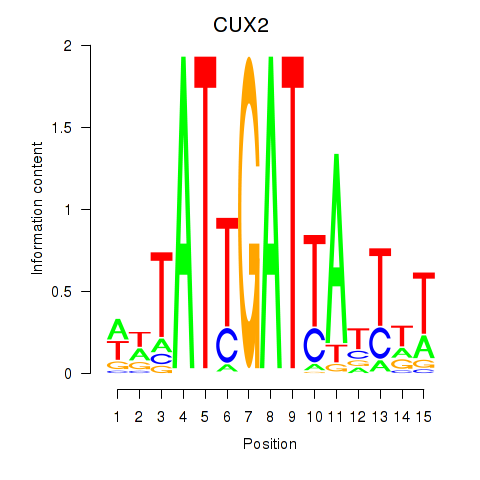
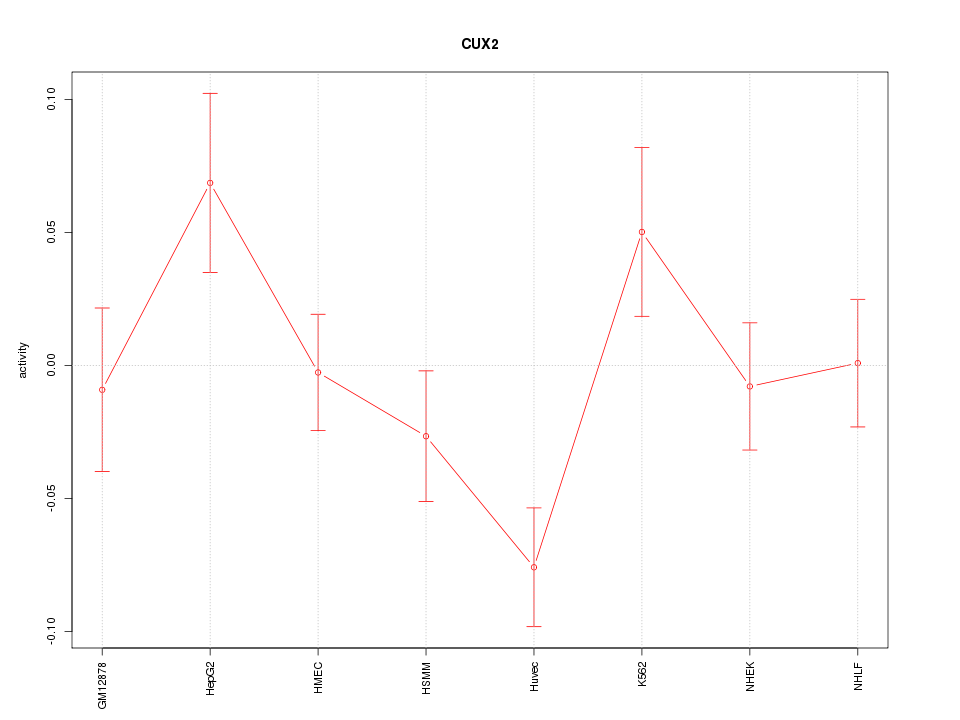
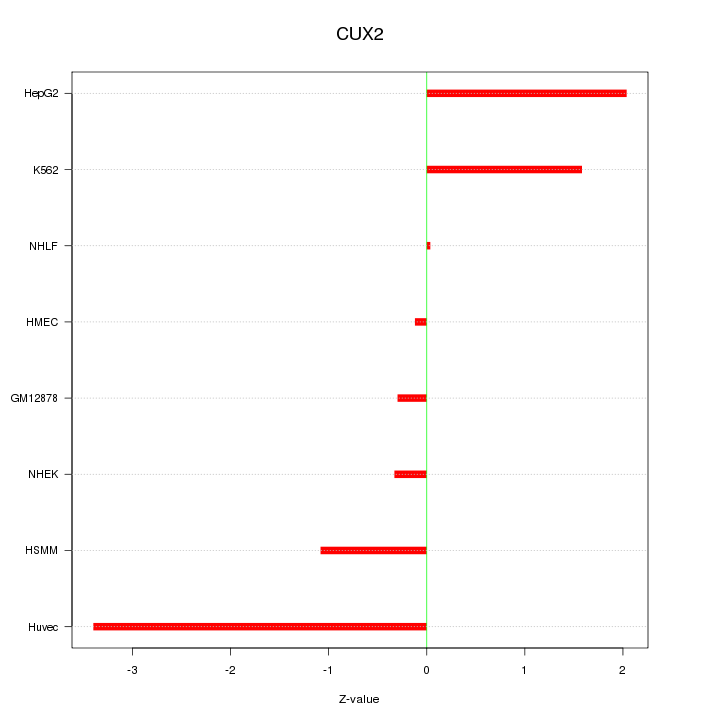
| 1.1 |
3.2 |
GO:0001907 |
cytolysis by symbiont of host cells(GO:0001897) killing by symbiont of host cells(GO:0001907) hemolysis by symbiont of host erythrocytes(GO:0019836) disruption by symbiont of host cell(GO:0044004) hemolysis in other organism(GO:0044179) positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) cytolysis in other organism(GO:0051715) cytolysis in other organism involved in symbiotic interaction(GO:0051801) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.9 |
8.7 |
GO:0051918 |
negative regulation of fibrinolysis(GO:0051918) |
| 0.8 |
2.3 |
GO:2000644 |
low-density lipoprotein particle receptor catabolic process(GO:0032802) regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) regulation of receptor catabolic process(GO:2000644) |
| 0.4 |
1.6 |
GO:0071400 |
cellular response to oleic acid(GO:0071400) |
| 0.4 |
1.2 |
GO:1903961 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.3 |
1.3 |
GO:0001705 |
ectoderm formation(GO:0001705) central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.3 |
1.4 |
GO:0032509 |
endosome transport via multivesicular body sorting pathway(GO:0032509) plasma membrane to endosome transport(GO:0048227) |
| 0.2 |
1.7 |
GO:0070779 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 |
0.6 |
GO:0002378 |
immunoglobulin biosynthetic process(GO:0002378) |
| 0.2 |
0.5 |
GO:0043095 |
regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.2 |
0.5 |
GO:0051919 |
positive regulation of fibrinolysis(GO:0051919) |
| 0.2 |
0.6 |
GO:0047484 |
regulation of response to osmotic stress(GO:0047484) |
| 0.2 |
1.1 |
GO:0006572 |
tyrosine catabolic process(GO:0006572) |
| 0.1 |
0.9 |
GO:0006537 |
glutamate biosynthetic process(GO:0006537) proline catabolic process(GO:0006562) |
| 0.1 |
0.4 |
GO:0072205 |
collecting duct development(GO:0072044) metanephric collecting duct development(GO:0072205) |
| 0.1 |
0.7 |
GO:0030210 |
heparin biosynthetic process(GO:0030210) |
| 0.1 |
0.9 |
GO:0051895 |
negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 |
5.8 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.1 |
0.5 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.1 |
0.3 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 |
1.7 |
GO:0006700 |
C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 |
0.3 |
GO:0021836 |
chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.1 |
1.9 |
GO:0000380 |
alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 |
0.5 |
GO:0051790 |
short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 |
2.3 |
GO:0034035 |
purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.1 |
0.5 |
GO:0007500 |
mesodermal cell fate determination(GO:0007500) |
| 0.1 |
0.2 |
GO:0072201 |
negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.1 |
0.5 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.1 |
0.4 |
GO:0060242 |
contact inhibition(GO:0060242) |
| 0.1 |
1.0 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 |
0.3 |
GO:0042519 |
tyrosine phosphorylation of Stat4 protein(GO:0042504) regulation of tyrosine phosphorylation of Stat4 protein(GO:0042519) positive regulation of tyrosine phosphorylation of Stat4 protein(GO:0042520) |
| 0.1 |
0.2 |
GO:0048852 |
diencephalon morphogenesis(GO:0048852) |
| 0.1 |
0.4 |
GO:0015742 |
alpha-ketoglutarate transport(GO:0015742) |
| 0.1 |
1.9 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.1 |
0.4 |
GO:0048296 |
isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 |
0.1 |
GO:0010430 |
ethanol catabolic process(GO:0006068) fatty acid omega-oxidation(GO:0010430) |
| 0.0 |
1.0 |
GO:0015804 |
neutral amino acid transport(GO:0015804) |
| 0.0 |
0.3 |
GO:0048853 |
forebrain morphogenesis(GO:0048853) |
| 0.0 |
0.1 |
GO:0034723 |
DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 |
0.4 |
GO:0034382 |
chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 |
0.1 |
GO:0010881 |
regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) |
| 0.0 |
0.1 |
GO:0044240 |
multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 |
0.2 |
GO:0048537 |
mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 |
0.1 |
GO:0016554 |
cytidine to uridine editing(GO:0016554) |
| 0.0 |
0.4 |
GO:0006776 |
vitamin A metabolic process(GO:0006776) |
| 0.0 |
0.2 |
GO:0045896 |
regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 |
0.4 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.0 |
0.1 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.0 |
0.1 |
GO:0010046 |
response to mycotoxin(GO:0010046) |
| 0.0 |
0.5 |
GO:0008210 |
estrogen metabolic process(GO:0008210) |
| 0.0 |
0.1 |
GO:0006422 |
aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 |
0.2 |
GO:0046051 |
GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.0 |
0.3 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.2 |
GO:0007598 |
blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 |
0.5 |
GO:0008209 |
androgen metabolic process(GO:0008209) |
| 0.0 |
0.1 |
GO:0030490 |
maturation of SSU-rRNA(GO:0030490) |
| 0.0 |
0.1 |
GO:0070245 |
positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 |
0.3 |
GO:0007128 |
meiotic prophase I(GO:0007128) |
| 0.0 |
0.1 |
GO:0002860 |
natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 |
0.2 |
GO:0019321 |
pentose metabolic process(GO:0019321) |
| 0.0 |
0.7 |
GO:0060338 |
regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 |
0.4 |
GO:0000038 |
very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 |
0.1 |
GO:0048147 |
negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 |
0.3 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.0 |
0.1 |
GO:0061025 |
membrane fusion(GO:0061025) |
| 0.0 |
0.4 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.0 |
0.7 |
GO:0002286 |
T cell activation involved in immune response(GO:0002286) |
| 0.0 |
0.1 |
GO:0060992 |
response to fungicide(GO:0060992) |
| 0.0 |
0.1 |
GO:0019934 |
cGMP-mediated signaling(GO:0019934) |
| 0.0 |
0.5 |
GO:0030866 |
cortical cytoskeleton organization(GO:0030865) cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 |
0.2 |
GO:0006400 |
tRNA modification(GO:0006400) |
| 0.0 |
1.1 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.0 |
0.1 |
GO:0002888 |
positive regulation of myeloid leukocyte mediated immunity(GO:0002888) positive regulation of mast cell activation(GO:0033005) positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 |
0.4 |
GO:0051925 |
obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.0 |
0.4 |
GO:0051930 |
regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 |
0.4 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.2 |
GO:0035338 |
long-chain fatty-acyl-CoA metabolic process(GO:0035336) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 |
1.0 |
GO:0008360 |
regulation of cell shape(GO:0008360) |
| 0.0 |
0.3 |
GO:0007172 |
signal complex assembly(GO:0007172) |
| 0.0 |
0.3 |
GO:0033209 |
tumor necrosis factor-mediated signaling pathway(GO:0033209) |