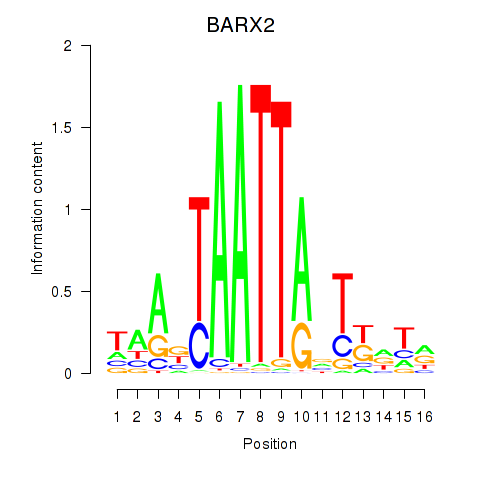
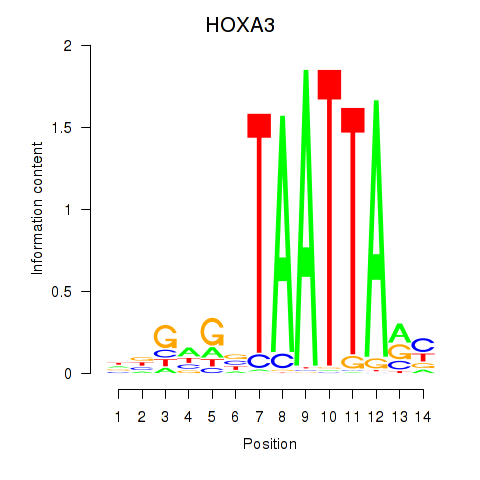
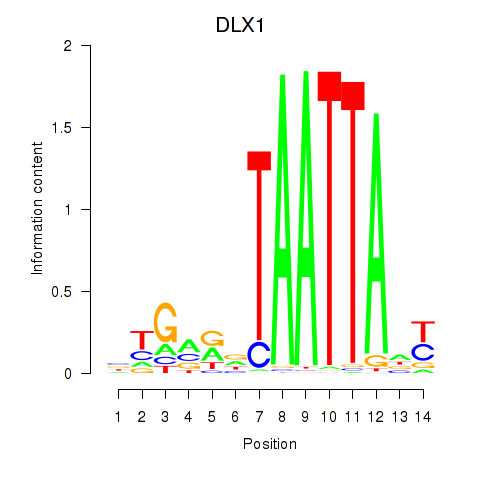
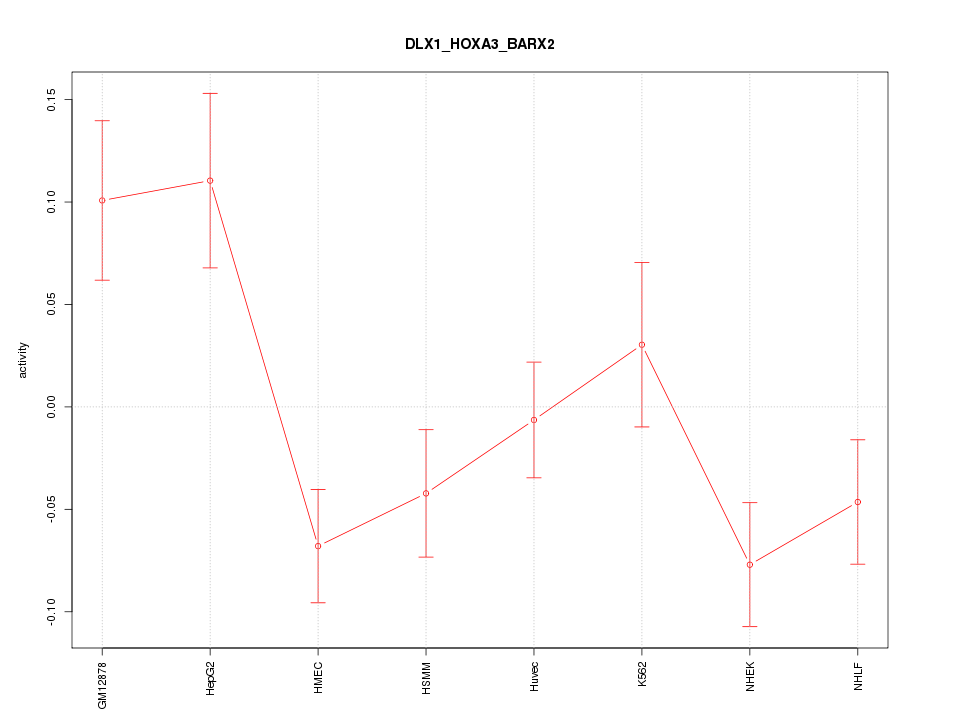
| 0.7 |
2.0 |
GO:1901419 |
regulation of response to alcohol(GO:1901419) |
| 0.6 |
1.9 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.6 |
1.8 |
GO:1904358 |
positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.5 |
2.2 |
GO:0010900 |
regulation of phosphatidylcholine catabolic process(GO:0010899) negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.5 |
2.7 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.5 |
1.5 |
GO:0001907 |
cytolysis by symbiont of host cells(GO:0001897) killing by symbiont of host cells(GO:0001907) hemolysis by symbiont of host erythrocytes(GO:0019836) disruption by symbiont of host cell(GO:0044004) hemolysis in other organism(GO:0044179) cytolysis in other organism(GO:0051715) cytolysis in other organism involved in symbiotic interaction(GO:0051801) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.4 |
1.2 |
GO:1903961 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.4 |
1.2 |
GO:0002378 |
immunoglobulin biosynthetic process(GO:0002378) |
| 0.4 |
1.9 |
GO:0010957 |
negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.4 |
1.5 |
GO:0018352 |
protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.4 |
2.9 |
GO:0034392 |
negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.3 |
0.9 |
GO:0051661 |
maintenance of centrosome location(GO:0051661) |
| 0.3 |
3.3 |
GO:0006975 |
DNA damage induced protein phosphorylation(GO:0006975) |
| 0.3 |
0.9 |
GO:0002431 |
Fc receptor mediated stimulatory signaling pathway(GO:0002431) regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.3 |
7.1 |
GO:0051923 |
purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) sulfation(GO:0051923) |
| 0.3 |
1.4 |
GO:0007095 |
blastocyst growth(GO:0001832) mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.3 |
0.8 |
GO:0010430 |
ethanol catabolic process(GO:0006068) fatty acid omega-oxidation(GO:0010430) |
| 0.3 |
0.8 |
GO:0042097 |
lymphotoxin A production(GO:0032641) interleukin-4 biosynthetic process(GO:0042097) lymphotoxin A biosynthetic process(GO:0042109) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.3 |
0.8 |
GO:0002752 |
cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.3 |
1.4 |
GO:0060164 |
regulation of timing of neuron differentiation(GO:0060164) |
| 0.3 |
0.8 |
GO:0048389 |
intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) bud dilation involved in lung branching(GO:0060503) branch elongation involved in ureteric bud branching(GO:0060681) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) pulmonary artery morphogenesis(GO:0061156) cell proliferation involved in mesonephros development(GO:0061209) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) negative regulation of mesonephros development(GO:0061218) pattern specification involved in mesonephros development(GO:0061227) renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) BMP signaling pathway involved in nephric duct formation(GO:0071893) glomerular visceral epithelial cell development(GO:0072015) comma-shaped body morphogenesis(GO:0072049) regulation of branch elongation involved in ureteric bud branching(GO:0072095) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) glomerulus morphogenesis(GO:0072102) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) nephric duct formation(GO:0072179) ureter urothelium development(GO:0072190) ureter epithelial cell differentiation(GO:0072192) ureter morphogenesis(GO:0072197) metanephric comma-shaped body morphogenesis(GO:0072278) glomerular epithelial cell development(GO:0072310) negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.2 |
1.2 |
GO:0007614 |
short-term memory(GO:0007614) |
| 0.2 |
0.7 |
GO:0043095 |
regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.2 |
5.6 |
GO:0002504 |
antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.2 |
0.9 |
GO:0070527 |
integrin activation(GO:0033622) regulation of integrin activation(GO:0033623) positive regulation of integrin activation(GO:0033625) platelet aggregation(GO:0070527) |
| 0.2 |
0.2 |
GO:0010891 |
negative regulation of lipid storage(GO:0010888) regulation of sequestering of triglyceride(GO:0010889) negative regulation of sequestering of triglyceride(GO:0010891) sequestering of triglyceride(GO:0030730) |
| 0.2 |
2.1 |
GO:0046653 |
tetrahydrofolate metabolic process(GO:0046653) |
| 0.2 |
1.0 |
GO:0046642 |
negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.2 |
0.8 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 |
2.7 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.2 |
0.2 |
GO:0071460 |
cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 |
2.5 |
GO:0046697 |
decidualization(GO:0046697) |
| 0.2 |
4.9 |
GO:0051225 |
spindle assembly(GO:0051225) |
| 0.2 |
2.7 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.2 |
0.6 |
GO:0048619 |
embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 |
1.0 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.1 |
0.4 |
GO:0010273 |
regulation of oxidative phosphorylation(GO:0002082) detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 |
0.4 |
GO:0002538 |
arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.1 |
0.5 |
GO:0010727 |
negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 |
0.4 |
GO:0006982 |
response to lipid hydroperoxide(GO:0006982) |
| 0.1 |
0.7 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 |
0.3 |
GO:0010172 |
embryonic body morphogenesis(GO:0010172) |
| 0.1 |
1.1 |
GO:0048013 |
ephrin receptor signaling pathway(GO:0048013) |
| 0.1 |
1.1 |
GO:0006979 |
response to oxidative stress(GO:0006979) |
| 0.1 |
1.1 |
GO:0050858 |
negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 |
2.6 |
GO:0033209 |
tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 |
0.3 |
GO:0008628 |
hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.1 |
1.1 |
GO:0071345 |
cellular response to cytokine stimulus(GO:0071345) |
| 0.1 |
0.4 |
GO:0032959 |
inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.1 |
0.9 |
GO:0042178 |
xenobiotic catabolic process(GO:0042178) |
| 0.1 |
0.4 |
GO:0050955 |
thermoception(GO:0050955) |
| 0.1 |
0.9 |
GO:0032515 |
negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 |
0.4 |
GO:0006562 |
proline catabolic process(GO:0006562) |
| 0.1 |
0.2 |
GO:0006435 |
threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 |
0.5 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.1 |
1.2 |
GO:0007342 |
fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 |
1.0 |
GO:0051319 |
mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.1 |
0.7 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.1 |
0.5 |
GO:0051383 |
kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.1 |
0.4 |
GO:0061099 |
negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 |
0.2 |
GO:0051971 |
positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.1 |
0.6 |
GO:0007351 |
tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.1 |
0.3 |
GO:0035444 |
nickel cation transport(GO:0015675) vanadium ion transport(GO:0015676) ferrous iron transport(GO:0015684) lead ion transport(GO:0015692) nickel cation transmembrane transport(GO:0035444) ferrous iron import(GO:0070627) iron ion import(GO:0097286) |
| 0.1 |
0.8 |
GO:0045087 |
innate immune response(GO:0045087) |
| 0.1 |
0.2 |
GO:0071436 |
sodium ion export(GO:0071436) |
| 0.1 |
1.4 |
GO:0008211 |
glucocorticoid metabolic process(GO:0008211) |
| 0.1 |
0.3 |
GO:0006515 |
misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 |
0.2 |
GO:0051823 |
hindbrain radial glia guided cell migration(GO:0021932) regulation of synapse structural plasticity(GO:0051823) |
| 0.1 |
0.5 |
GO:0030299 |
intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.1 |
0.4 |
GO:0051260 |
protein homooligomerization(GO:0051260) |
| 0.0 |
0.2 |
GO:0050857 |
positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 |
0.9 |
GO:0006359 |
regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 |
0.1 |
GO:1904396 |
regulation of synaptic growth at neuromuscular junction(GO:0008582) regulation of neuromuscular junction development(GO:1904396) |
| 0.0 |
1.6 |
GO:0042475 |
odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 |
0.2 |
GO:0042519 |
tyrosine phosphorylation of Stat4 protein(GO:0042504) regulation of tyrosine phosphorylation of Stat4 protein(GO:0042519) positive regulation of tyrosine phosphorylation of Stat4 protein(GO:0042520) |
| 0.0 |
2.9 |
GO:0019882 |
antigen processing and presentation(GO:0019882) |
| 0.0 |
0.4 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
0.2 |
GO:0006538 |
glutamate catabolic process(GO:0006538) |
| 0.0 |
0.2 |
GO:0010890 |
positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 |
0.6 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.0 |
0.6 |
GO:0090399 |
replicative senescence(GO:0090399) |
| 0.0 |
2.2 |
GO:0008206 |
bile acid metabolic process(GO:0008206) |
| 0.0 |
0.2 |
GO:0021759 |
globus pallidus development(GO:0021759) menarche(GO:0042696) |
| 0.0 |
1.3 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.0 |
0.2 |
GO:0000239 |
pachytene(GO:0000239) |
| 0.0 |
0.4 |
GO:0016998 |
cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 |
0.1 |
GO:1901642 |
purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.0 |
0.8 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.0 |
0.3 |
GO:0033089 |
positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 |
0.4 |
GO:0009615 |
response to virus(GO:0009615) |
| 0.0 |
0.5 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 |
0.2 |
GO:0001507 |
acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 |
0.3 |
GO:0009750 |
response to fructose(GO:0009750) |
| 0.0 |
0.5 |
GO:0034332 |
adherens junction organization(GO:0034332) |
| 0.0 |
0.1 |
GO:0001692 |
histamine metabolic process(GO:0001692) |
| 0.0 |
0.2 |
GO:0021516 |
dorsal spinal cord development(GO:0021516) |
| 0.0 |
0.3 |
GO:0030147 |
obsolete natriuresis(GO:0030147) |
| 0.0 |
1.0 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.4 |
GO:0016601 |
Rac protein signal transduction(GO:0016601) |
| 0.0 |
0.1 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.0 |
0.1 |
GO:0006668 |
sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 |
1.0 |
GO:0060338 |
regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 |
1.5 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
0.7 |
GO:0071804 |
cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 |
2.3 |
GO:0006096 |
glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 |
0.1 |
GO:0048537 |
mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 |
2.5 |
GO:0000236 |
mitotic prometaphase(GO:0000236) |
| 0.0 |
0.4 |
GO:0014046 |
dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 |
0.2 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.2 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.0 |
0.2 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 |
0.6 |
GO:0051925 |
obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.0 |
0.0 |
GO:0002331 |
immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.0 |
0.8 |
GO:0046324 |
regulation of glucose import(GO:0046324) |
| 0.0 |
1.6 |
GO:0006959 |
humoral immune response(GO:0006959) |
| 0.0 |
0.1 |
GO:0009698 |
phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.0 |
0.7 |
GO:0015758 |
hexose transport(GO:0008645) glucose transport(GO:0015758) |
| 0.0 |
0.6 |
GO:0043154 |
negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 |
0.3 |
GO:0051568 |
histone H3-K4 methylation(GO:0051568) |
| 0.0 |
0.3 |
GO:0045749 |
obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 |
2.2 |
GO:0008203 |
cholesterol metabolic process(GO:0008203) |
| 0.0 |
0.9 |
GO:0006694 |
steroid biosynthetic process(GO:0006694) |
| 0.0 |
0.1 |
GO:0060968 |
regulation of gene silencing(GO:0060968) |
| 0.0 |
0.2 |
GO:0071044 |
histone mRNA catabolic process(GO:0071044) |
| 0.0 |
0.3 |
GO:0048260 |
positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 |
0.6 |
GO:0042771 |
intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 |
0.0 |
GO:0016098 |
monoterpenoid metabolic process(GO:0016098) |
| 0.0 |
0.4 |
GO:0000060 |
protein import into nucleus, translocation(GO:0000060) |
| 0.0 |
0.2 |
GO:0045123 |
cellular extravasation(GO:0045123) |
| 0.0 |
1.1 |
GO:0031295 |
lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 |
0.1 |
GO:1900087 |
traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.0 |
0.2 |
GO:0090630 |
activation of GTPase activity(GO:0090630) |
| 0.0 |
0.3 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 |
0.1 |
GO:0043438 |
acetoacetic acid metabolic process(GO:0043438) |
| 0.0 |
3.1 |
GO:0097190 |
apoptotic signaling pathway(GO:0097190) |
| 0.0 |
0.5 |
GO:0043524 |
negative regulation of neuron apoptotic process(GO:0043524) negative regulation of neuron death(GO:1901215) |
| 0.0 |
0.1 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.0 |
0.2 |
GO:0008334 |
histone mRNA metabolic process(GO:0008334) |
| 0.0 |
2.0 |
GO:0050658 |
nucleic acid transport(GO:0050657) RNA transport(GO:0050658) establishment of RNA localization(GO:0051236) |
| 0.0 |
0.2 |
GO:0015695 |
organic cation transport(GO:0015695) |
| 0.0 |
0.3 |
GO:0007274 |
neuromuscular synaptic transmission(GO:0007274) |
| 0.0 |
0.1 |
GO:0048535 |
lymph node development(GO:0048535) |
| 0.0 |
0.2 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 |
0.2 |
GO:0009395 |
phospholipid catabolic process(GO:0009395) |
| 0.0 |
4.5 |
GO:0051301 |
cell division(GO:0051301) |
| 0.0 |
0.1 |
GO:0050892 |
intestinal absorption(GO:0050892) |
| 0.0 |
0.1 |
GO:0006706 |
steroid catabolic process(GO:0006706) |
| 0.0 |
0.3 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |