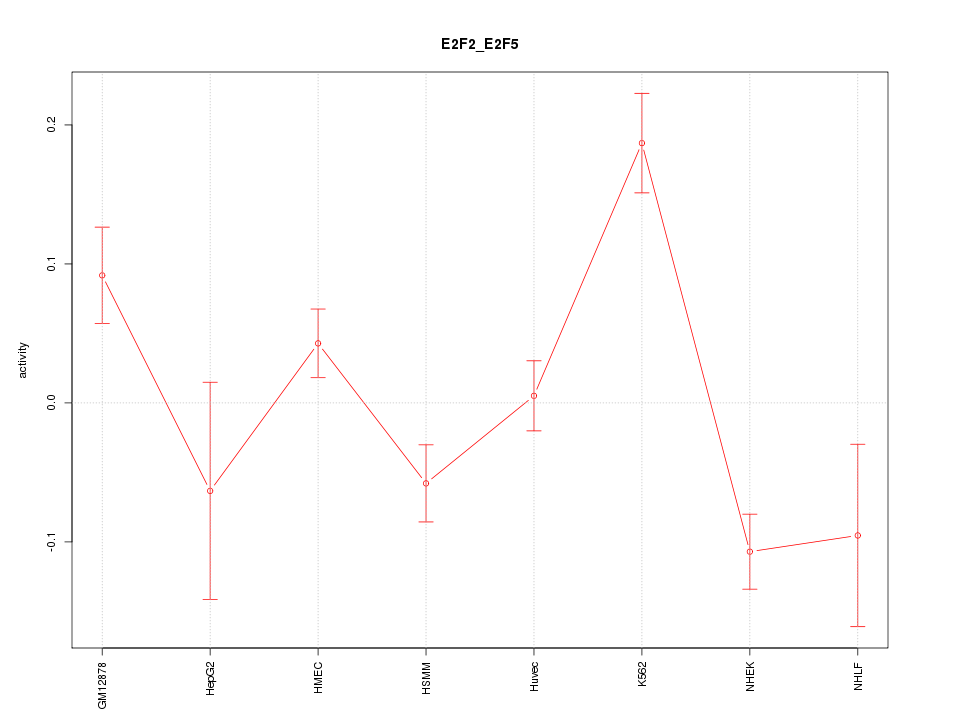
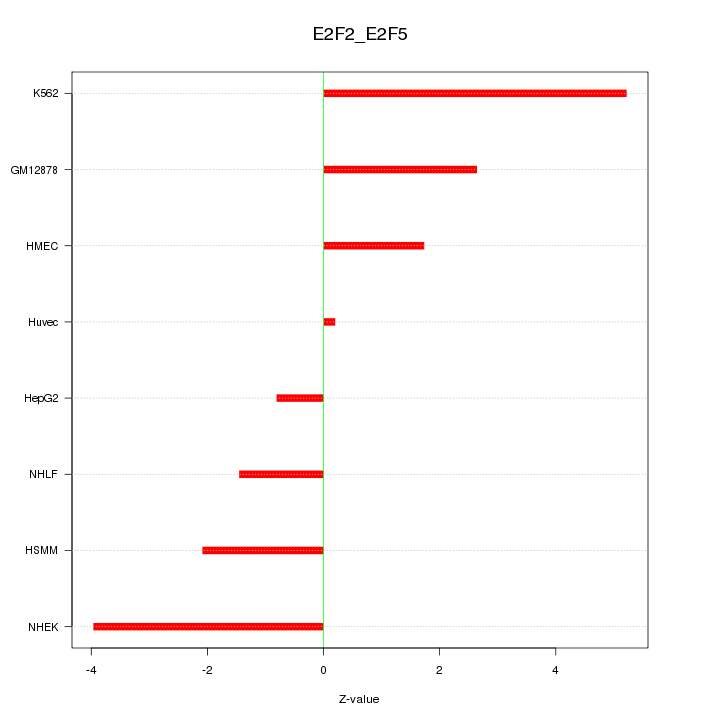
Motif ID: E2F2_E2F5
Z-value: 2.742
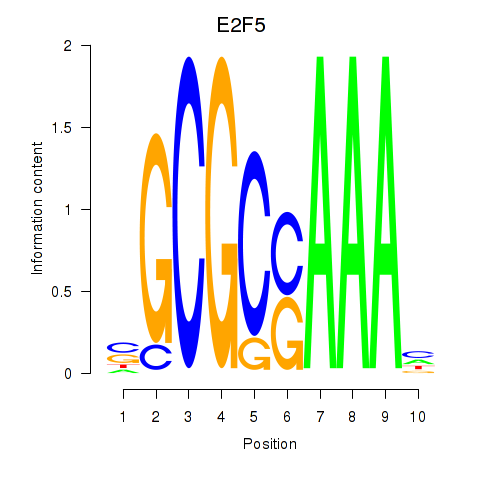
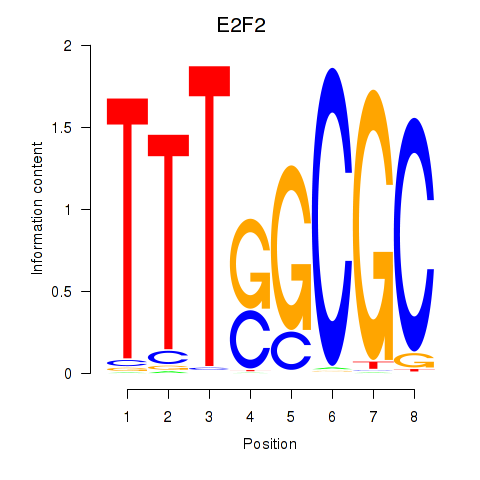
Transcription factors associated with E2F2_E2F5:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| E2F2 | ENSG00000007968.6 | E2F2 |
| E2F5 | ENSG00000133740.6 | E2F5 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 1.6 | 9.5 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 1.5 | 4.4 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 1.3 | 3.9 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 1.0 | 4.1 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 1.0 | 4.0 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.9 | 2.8 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.9 | 32.6 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.8 | 2.3 | GO:1901724 | positive regulation of cell proliferation involved in kidney development(GO:1901724) |
| 0.8 | 7.0 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.7 | 7.3 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.7 | 5.7 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.7 | 2.7 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.6 | 1.8 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.6 | 2.4 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.5 | 2.7 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.5 | 1.4 | GO:0046604 | regulation of mitotic centrosome separation(GO:0046602) positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.3 | 1.0 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.3 | 4.7 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.3 | 2.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.3 | 2.0 | GO:0033504 | floor plate development(GO:0033504) |
| 0.3 | 5.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.3 | 1.8 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.2 | 1.6 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 10.7 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.2 | 1.0 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.2 | 1.1 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.2 | 1.3 | GO:0031657 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.2 | 0.5 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 | 1.1 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.6 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.1 | 3.9 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 12.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.7 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) |
| 0.1 | 0.4 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.1 | 1.2 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 1.1 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.1 | 0.5 | GO:0000239 | pachytene(GO:0000239) |
| 0.1 | 0.4 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 1.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 1.0 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) |
| 0.1 | 1.9 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.1 | 0.4 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.1 | 3.4 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.1 | 0.3 | GO:0072387 | flavin-containing compound biosynthetic process(GO:0042727) flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.1 | 0.4 | GO:0060394 | obsolete positive regulation of S phase of mitotic cell cycle(GO:0045750) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.6 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.1 | 7.8 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 0.4 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.1 | 2.6 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 0.8 | GO:0051567 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.1 | 0.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 1.7 | GO:0006260 | DNA replication(GO:0006260) |
| 0.1 | 0.6 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 0.8 | GO:0071398 | cellular response to fatty acid(GO:0071398) |
| 0.1 | 0.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 1.7 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.1 | 0.5 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.1 | 0.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.5 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 0.9 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 1.4 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 0.7 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 1.2 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 0.4 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.6 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.4 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 1.3 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 2.0 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.8 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.2 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 2.8 | GO:0000216 | obsolete M/G1 transition of mitotic cell cycle(GO:0000216) |
| 0.0 | 0.4 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 11.0 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 1.6 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 1.0 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.6 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.6 | GO:0000279 | M phase(GO:0000279) |
| 0.0 | 0.3 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.0 | 0.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 1.6 | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.7 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.3 | GO:0007628 | adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.0 | 1.5 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.3 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 0.9 | GO:0044843 | G1/S transition of mitotic cell cycle(GO:0000082) cell cycle G1/S phase transition(GO:0044843) |
| 0.0 | 0.1 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.2 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.4 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 25.1 | GO:0042555 | MCM complex(GO:0042555) |
| 1.4 | 5.7 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 1.2 | 4.7 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.9 | 0.9 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.7 | 9.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.6 | 9.6 | GO:0030894 | replisome(GO:0030894) nuclear replisome(GO:0043601) |
| 0.5 | 1.5 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.3 | 2.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.3 | 0.9 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.3 | 2.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.3 | 1.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.3 | 13.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 1.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 5.2 | GO:0005657 | replication fork(GO:0005657) |
| 0.2 | 1.9 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.2 | 0.8 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.2 | 9.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.2 | 0.8 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 11.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 1.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 2.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.6 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 0.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 2.1 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.5 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 1.1 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.0 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 5.1 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.4 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.5 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.4 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 1.1 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 10.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.8 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 26.1 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 1.2 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.9 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.8 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.9 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.4 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.0 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 1.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0004146 | dihydrofolate reductase activity(GO:0004146) |
| 1.2 | 4.7 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.9 | 0.9 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.9 | 2.8 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.9 | 3.5 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.6 | 0.6 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.6 | 4.2 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.4 | 9.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.4 | 3.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.3 | 12.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.3 | 1.0 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.3 | 0.6 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.3 | 0.8 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.3 | 7.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.3 | 5.5 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.2 | 0.7 | GO:0030611 | arsenate reductase activity(GO:0030611) |
| 0.2 | 0.7 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.2 | 1.1 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.2 | 6.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 2.3 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.2 | 7.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.2 | 1.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 0.9 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 3.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 5.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 6.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 12.0 | GO:0016566 | obsolete specific transcriptional repressor activity(GO:0016566) |
| 0.1 | 0.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.5 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 1.1 | GO:0045502 | dynein binding(GO:0045502) |
| 0.1 | 1.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.4 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.1 | 0.3 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 1.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.6 | GO:0016174 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.6 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.4 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 1.0 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.0 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 13.0 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.4 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.3 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 4.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.1 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 1.2 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.4 | GO:0001104 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 1.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 28.7 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 3.9 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 0.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 1.3 | GO:0015631 | tubulin binding(GO:0015631) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.5 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.8 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |