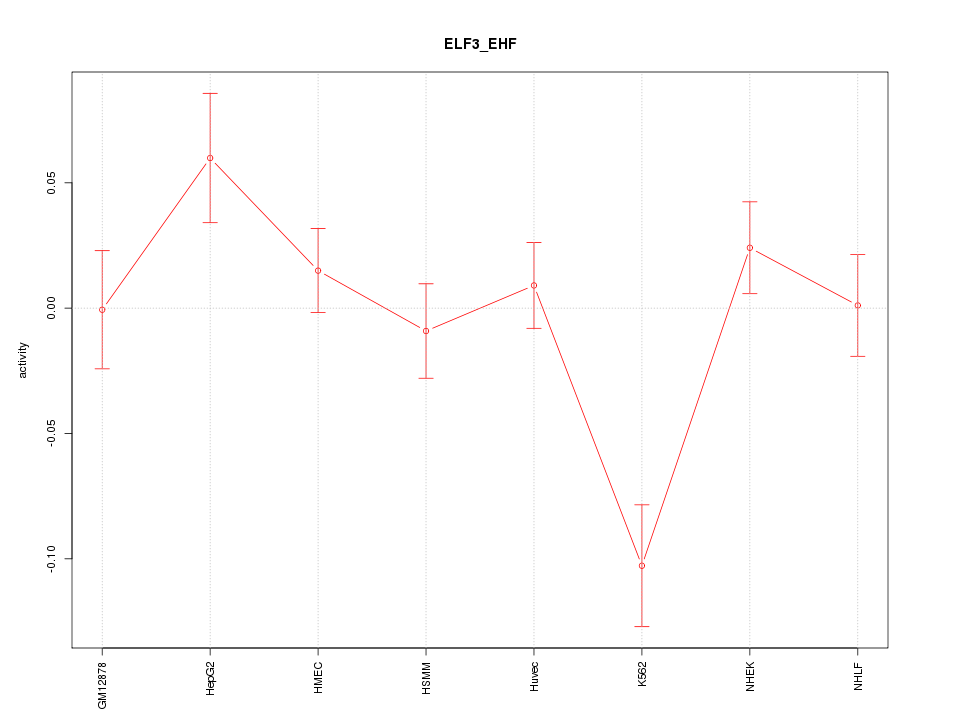
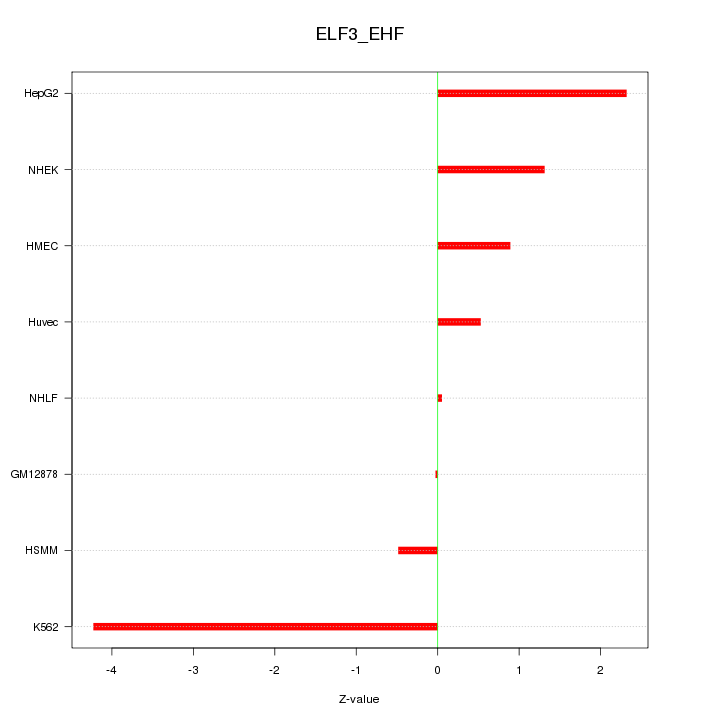
Motif ID: ELF3_EHF
Z-value: 1.813
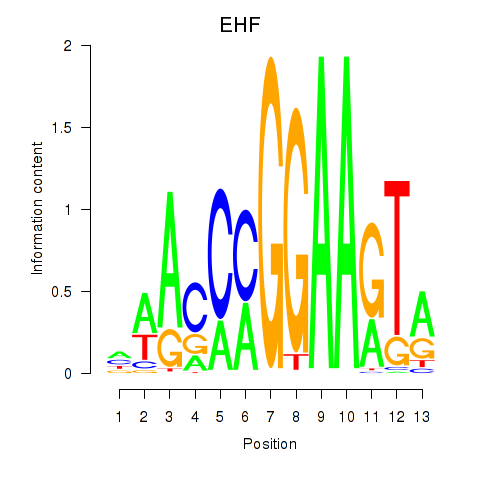
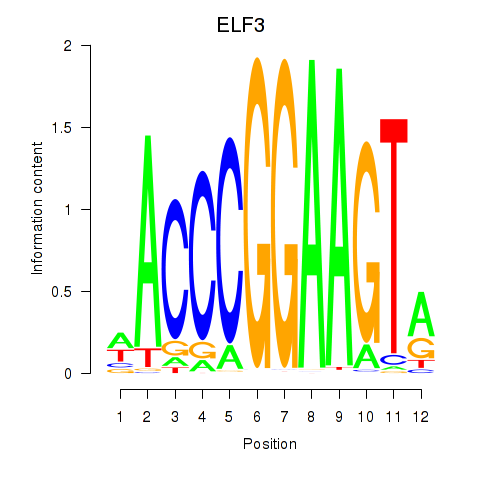
Transcription factors associated with ELF3_EHF:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| EHF | ENSG00000135373.8 | EHF |
| ELF3 | ENSG00000163435.11 | ELF3 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.8 | 3.3 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.8 | 2.5 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.6 | 1.9 | GO:1903961 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.6 | 2.3 | GO:0034442 | plasma lipoprotein particle oxidation(GO:0034441) regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) lipoprotein modification(GO:0042160) lipoprotein oxidation(GO:0042161) |
| 0.4 | 1.3 | GO:0010757 | chronological cell aging(GO:0001300) arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) negative regulation of plasminogen activation(GO:0010757) negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.4 | 5.4 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.3 | 8.5 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.3 | 1.0 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.3 | 0.8 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) negative regulation of protein glycosylation(GO:0060051) |
| 0.3 | 1.0 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.2 | 1.3 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.2 | 0.9 | GO:0032870 | cellular response to hormone stimulus(GO:0032870) |
| 0.2 | 1.4 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.2 | 0.8 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.2 | 0.2 | GO:2001021 | negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.2 | 0.7 | GO:0060209 | estrus(GO:0060209) |
| 0.2 | 0.7 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.2 | 0.8 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.2 | 1.2 | GO:2000109 | macrophage apoptotic process(GO:0071888) regulation of macrophage apoptotic process(GO:2000109) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.2 | 2.3 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.2 | 0.5 | GO:0034244 | forebrain anterior/posterior pattern specification(GO:0021797) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) neural plate anterior/posterior regionalization(GO:0021999) negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) axial mesoderm morphogenesis(GO:0048319) neural plate pattern specification(GO:0060896) neural plate regionalization(GO:0060897) |
| 0.2 | 0.5 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.2 | 1.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 1.2 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 2.2 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.1 | 0.7 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.3 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 0.4 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.1 | 0.7 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.5 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.4 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.1 | 0.5 | GO:0046121 | deoxyribonucleoside metabolic process(GO:0009120) deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.4 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 0.4 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 1.0 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.1 | 2.8 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.1 | 0.8 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.7 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.1 | 0.3 | GO:0003289 | septum primum development(GO:0003284) atrial septum primum morphogenesis(GO:0003289) |
| 0.1 | 0.7 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.3 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.1 | 0.3 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 0.4 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.1 | 0.4 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.1 | 0.8 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 0.7 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 1.3 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.5 | GO:0001816 | cytokine production(GO:0001816) |
| 0.1 | 0.8 | GO:0097384 | cellular lipid biosynthetic process(GO:0097384) |
| 0.1 | 1.0 | GO:0046473 | phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 0.2 | GO:0071504 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.1 | 1.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.3 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.5 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.1 | 1.5 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.1 | 0.3 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 0.6 | GO:0060414 | aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.4 | GO:0097193 | intrinsic apoptotic signaling pathway(GO:0097193) |
| 0.1 | 0.3 | GO:1990170 | detoxification of cadmium ion(GO:0071585) stress response to cadmium ion(GO:1990170) |
| 0.1 | 0.6 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.1 | 0.2 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.1 | 0.2 | GO:1903519 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.1 | 0.5 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 1.3 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.1 | 0.5 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.6 | GO:0003065 | regulation of heart rate by chemical signal(GO:0003062) positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.8 | GO:0042981 | regulation of apoptotic process(GO:0042981) |
| 0.1 | 0.3 | GO:0071801 | podosome assembly(GO:0071800) regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.1 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.1 | 0.4 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 1.0 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.1 | 1.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.6 | GO:0018126 | protein hydroxylation(GO:0018126) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.3 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.1 | 1.0 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.1 | 0.4 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.2 | GO:0035269 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.1 | 0.3 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.3 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) actin filament reorganization(GO:0090527) |
| 0.1 | 0.1 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.1 | 0.4 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.5 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 0.4 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.2 | GO:0003351 | cilium movement(GO:0003341) epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.3 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.9 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.1 | 0.4 | GO:0006896 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.1 | 0.1 | GO:0045978 | negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.0 | 0.4 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.5 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0090042 | tubulin deacetylation(GO:0090042) |
| 0.0 | 1.0 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.1 | GO:0021529 | type B pancreatic cell development(GO:0003323) spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.2 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 1.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.4 | GO:0030030 | cell projection organization(GO:0030030) |
| 0.0 | 1.0 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.1 | GO:0042519 | tyrosine phosphorylation of Stat4 protein(GO:0042504) regulation of tyrosine phosphorylation of Stat4 protein(GO:0042519) positive regulation of tyrosine phosphorylation of Stat4 protein(GO:0042520) |
| 0.0 | 0.6 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.5 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 1.4 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 6.5 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.1 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.4 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.1 | GO:1901659 | nucleoside biosynthetic process(GO:0009163) ribonucleoside biosynthetic process(GO:0042455) glycosyl compound biosynthetic process(GO:1901659) |
| 0.0 | 0.5 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.2 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.4 | GO:1901984 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.1 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.7 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.4 | GO:0032613 | interleukin-10 production(GO:0032613) |
| 0.0 | 0.5 | GO:0070192 | synapsis(GO:0007129) chromosome organization involved in meiotic cell cycle(GO:0070192) |
| 0.0 | 1.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.5 | GO:0010458 | exit from mitosis(GO:0010458) |
| 0.0 | 0.1 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.2 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:1900087 | traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.0 | 0.8 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.2 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 1.1 | GO:0008633 | obsolete activation of pro-apoptotic gene products(GO:0008633) |
| 0.0 | 0.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.3 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.2 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 1.1 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.8 | GO:0051925 | obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.0 | 0.2 | GO:0050912 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.0 | 0.1 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 2.4 | GO:0006886 | intracellular protein transport(GO:0006886) |
| 0.0 | 0.3 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.0 | 0.4 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.0 | 1.1 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.3 | GO:0050718 | positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.0 | 0.4 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.1 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 1.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.3 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.5 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.1 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 1.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.4 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.1 | GO:1902803 | regulation of synaptic vesicle transport(GO:1902803) regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 1.5 | GO:0043281 | regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043281) |
| 0.0 | 0.1 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.2 | GO:0018198 | peptidyl-cysteine modification(GO:0018198) |
| 0.0 | 0.2 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) |
| 0.0 | 0.1 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.4 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 0.2 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.3 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.0 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.6 | GO:0051017 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.0 | 0.7 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.0 | 0.7 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.2 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.6 | GO:0006029 | proteoglycan metabolic process(GO:0006029) |
| 0.0 | 0.2 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.2 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.1 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.1 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 | 0.5 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 1.6 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.4 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 1.9 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.1 | GO:0036503 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) ERAD pathway(GO:0036503) |
| 0.0 | 0.1 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 1.0 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.6 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 0.4 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 1.0 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.9 | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.1 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.2 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:0010821 | regulation of mitochondrion organization(GO:0010821) |
| 0.0 | 0.6 | GO:0001889 | liver development(GO:0001889) |
| 0.0 | 0.6 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.0 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.6 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.0 | 2.1 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.3 | GO:0031424 | keratinization(GO:0031424) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.4 | 2.2 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.3 | 0.8 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.2 | 1.4 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.2 | 0.6 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 0.5 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.2 | 0.7 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 0.7 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 1.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 2.3 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 2.7 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 1.3 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.1 | 0.5 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 0.7 | GO:0043186 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.1 | 0.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 2.2 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 1.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.6 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.3 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.7 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.3 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.1 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.3 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.1 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.8 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.1 | 0.6 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 0.6 | GO:0032059 | bleb(GO:0032059) |
| 0.1 | 0.3 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.4 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 1.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 3.8 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.6 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 2.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 3.3 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.9 | GO:0099568 | cytoplasmic region(GO:0099568) |
| 0.0 | 2.5 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 5.2 | GO:0019717 | obsolete synaptosome(GO:0019717) |
| 0.0 | 2.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.5 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 2.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.6 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 1.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.5 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0045179 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) apical cortex(GO:0045179) |
| 0.0 | 3.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) organelle outer membrane(GO:0031968) |
| 0.0 | 1.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.6 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 4.2 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.1 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0097610 | cell division site(GO:0032153) cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 5.0 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.3 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.3 | GO:0001533 | cornified envelope(GO:0001533) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 8.5 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.8 | 2.5 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.7 | 2.0 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.5 | 3.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.5 | 1.4 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.4 | 3.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 0.9 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.3 | 3.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 1.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.2 | 3.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.2 | 1.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.2 | 0.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 0.6 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.2 | 0.8 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.2 | 0.8 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 0.7 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.2 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 0.7 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 0.5 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.2 | 1.4 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.2 | 0.5 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.2 | 1.2 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.2 | 0.5 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.2 | 0.5 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.2 | 0.6 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.1 | 0.7 | GO:0015520 | tetracycline:proton antiporter activity(GO:0015520) |
| 0.1 | 0.6 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 1.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.7 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.4 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.1 | 0.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.7 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.1 | 0.5 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.7 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 2.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.3 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.1 | 0.5 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.1 | 2.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.3 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.1 | 3.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.9 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 1.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 1.9 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 0.4 | GO:0004803 | transposase activity(GO:0004803) |
| 0.1 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.3 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.1 | 0.2 | GO:0016731 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 0.4 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 0.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.9 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.5 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.2 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.1 | 0.3 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 0.8 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.5 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 1.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 1.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.5 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 0.2 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 0.4 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 4.5 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 0.5 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.2 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 0.7 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.1 | GO:0009384 | N-acylmannosamine kinase activity(GO:0009384) |
| 0.1 | 0.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.3 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.1 | 1.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.4 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 1.4 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 1.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.2 | GO:0004461 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) lactose synthase activity(GO:0004461) |
| 0.0 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 1.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.5 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.8 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 1.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.3 | GO:0043028 | cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 2.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.0 | 1.2 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.0 | 0.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 2.0 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 0.4 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.2 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 1.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.0 | GO:0004221 | obsolete ubiquitin thiolesterase activity(GO:0004221) |
| 0.0 | 0.1 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.1 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.3 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.6 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 6.8 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.3 | GO:0043499 | obsolete eukaryotic cell surface binding(GO:0043499) |
| 0.0 | 0.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.1 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.2 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.2 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | SIG_PIP3_SIGNALING_IN_B_LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 2.2 | SA_TRKA_RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 2.7 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 0.2 | ST_PAC1_RECEPTOR_PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.2 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.9 | ST_GAQ_PATHWAY | G alpha q Pathway |
| 0.0 | 1.6 | SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.8 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.1 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |