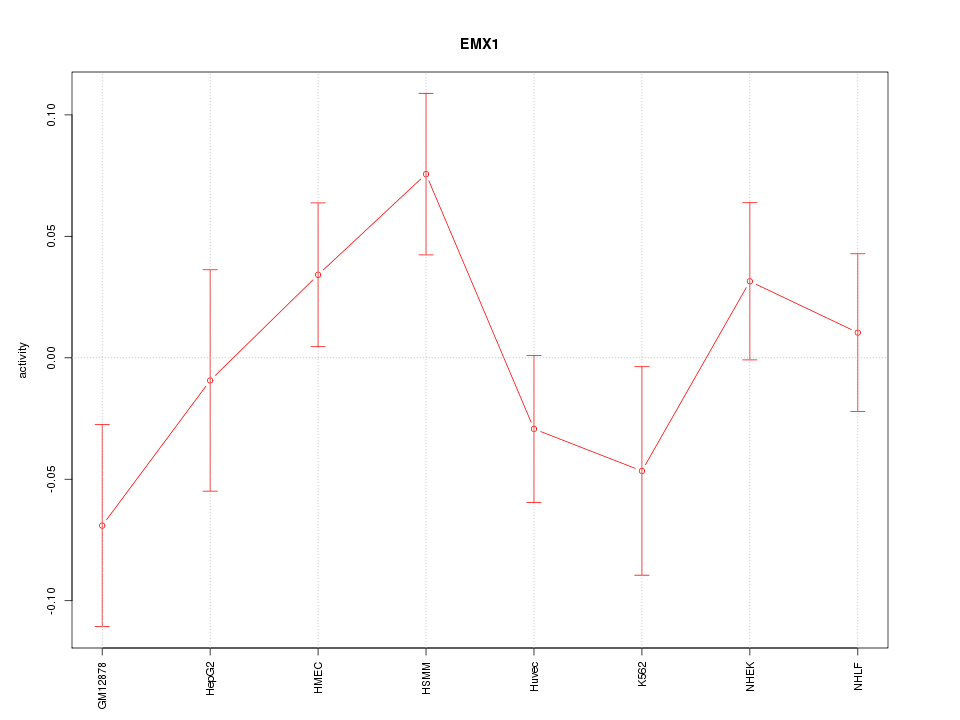
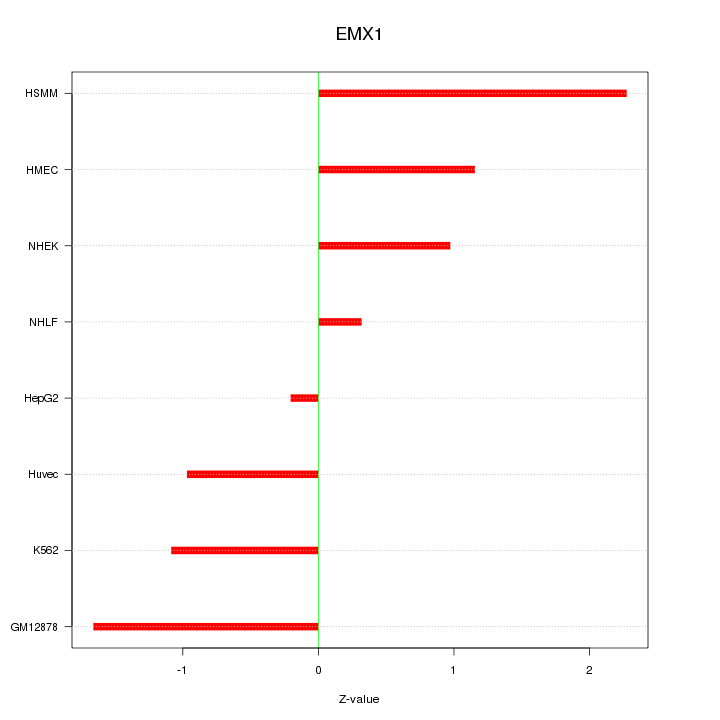
Motif ID: EMX1
Z-value: 1.248
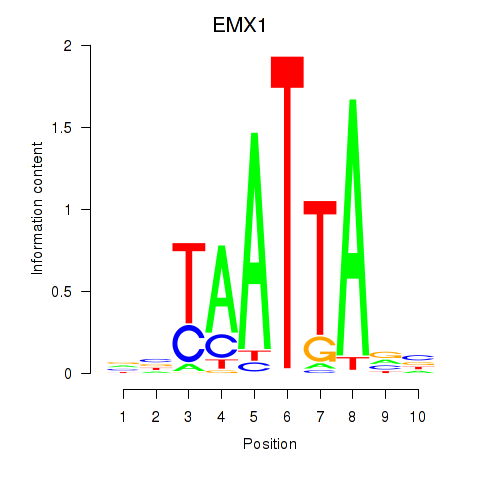
Transcription factors associated with EMX1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| EMX1 | ENSG00000135638.9 | EMX1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.5 | 2.0 | GO:0060649 | nipple development(GO:0060618) mammary gland bud elongation(GO:0060649) nipple morphogenesis(GO:0060658) nipple sheath formation(GO:0060659) |
| 0.4 | 1.2 | GO:0014889 | muscle atrophy(GO:0014889) regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.4 | 1.6 | GO:0031223 | auditory behavior(GO:0031223) malate-aspartate shuttle(GO:0043490) |
| 0.3 | 1.0 | GO:0031034 | skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 | 1.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.3 | 2.3 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.3 | 1.6 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.2 | 2.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 1.9 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.2 | 1.4 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.2 | 1.2 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.2 | 1.3 | GO:0060414 | aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 1.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.2 | 0.8 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 0.4 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 0.5 | GO:0071418 | uroporphyrinogen III biosynthetic process(GO:0006780) cellular response to amine stimulus(GO:0071418) |
| 0.1 | 1.4 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 1.2 | GO:0010658 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 0.6 | GO:0034204 | Cdc42 protein signal transduction(GO:0032488) lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 0.7 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 1.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.8 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.1 | 0.6 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) proline transport(GO:0015824) |
| 0.1 | 0.5 | GO:0010224 | response to UV-B(GO:0010224) |
| 0.1 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.2 | GO:0046520 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.5 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 1.8 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.1 | GO:0050942 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.1 | 0.5 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 1.5 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.1 | 1.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 4.5 | GO:0009408 | response to heat(GO:0009408) |
| 0.1 | 0.2 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.1 | 0.2 | GO:1902414 | protein localization to adherens junction(GO:0071896) protein localization to cell junction(GO:1902414) |
| 0.1 | 0.2 | GO:0070781 | response to biotin(GO:0070781) |
| 0.1 | 0.3 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.8 | GO:0042517 | positive regulation of tyrosine phosphorylation of Stat3 protein(GO:0042517) |
| 0.0 | 0.4 | GO:0048387 | protein maturation by protein folding(GO:0022417) negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 2.1 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.0 | 4.8 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 1.1 | GO:0006693 | prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:0046621 | negative regulation of organ growth(GO:0046621) |
| 0.0 | 0.7 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.3 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.0 | 0.9 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.6 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 2.8 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 0.2 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.7 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.4 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 0.4 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 2.1 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.4 | GO:0007128 | meiotic prophase I(GO:0007128) |
| 0.0 | 0.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) sulfur compound catabolic process(GO:0044273) |
| 0.0 | 0.5 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.1 | GO:0032472 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) Golgi calcium ion transport(GO:0032472) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.1 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.7 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 1.2 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.3 | GO:0001756 | somitogenesis(GO:0001756) somite development(GO:0061053) |
| 0.0 | 0.1 | GO:0034382 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) chylomicron remnant clearance(GO:0034382) phosphatidylcholine catabolic process(GO:0034638) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.0 | 0.1 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 | 0.3 | GO:0036503 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) ERAD pathway(GO:0036503) |
| 0.0 | 0.0 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 6.6 | GO:0007010 | cytoskeleton organization(GO:0007010) |
| 0.0 | 1.2 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.6 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) actin-mediated cell contraction(GO:0070252) |
| 0.0 | 0.3 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.2 | 2.5 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 1.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 1.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.7 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 4.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 1.6 | GO:0043205 | fibril(GO:0043205) |
| 0.1 | 0.3 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.1 | 1.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 2.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.5 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.4 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 1.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 2.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.1 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.5 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.8 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 3.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.2 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 1.0 | 5.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.3 | 1.1 | GO:0047026 | androsterone dehydrogenase (A-specific) activity(GO:0047026) |
| 0.3 | 1.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 1.0 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.2 | 0.4 | GO:0047718 | trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) indanol dehydrogenase activity(GO:0047718) |
| 0.2 | 1.8 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.1 | 1.4 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.6 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 1.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 3.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.5 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 5.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 1.2 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 1.1 | GO:0016944 | obsolete RNA polymerase II transcription elongation factor activity(GO:0016944) |
| 0.1 | 2.1 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 1.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.5 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.1 | 0.8 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.2 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 0.7 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.1 | 0.5 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 1.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 1.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.4 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.6 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 2.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.1 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 0.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 1.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.0 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 1.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 2.7 | GO:0003702 | obsolete RNA polymerase II transcription factor activity(GO:0003702) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 1.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.0 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | SIG_IL4RECEPTOR_IN_B_LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 1.2 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.2 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |