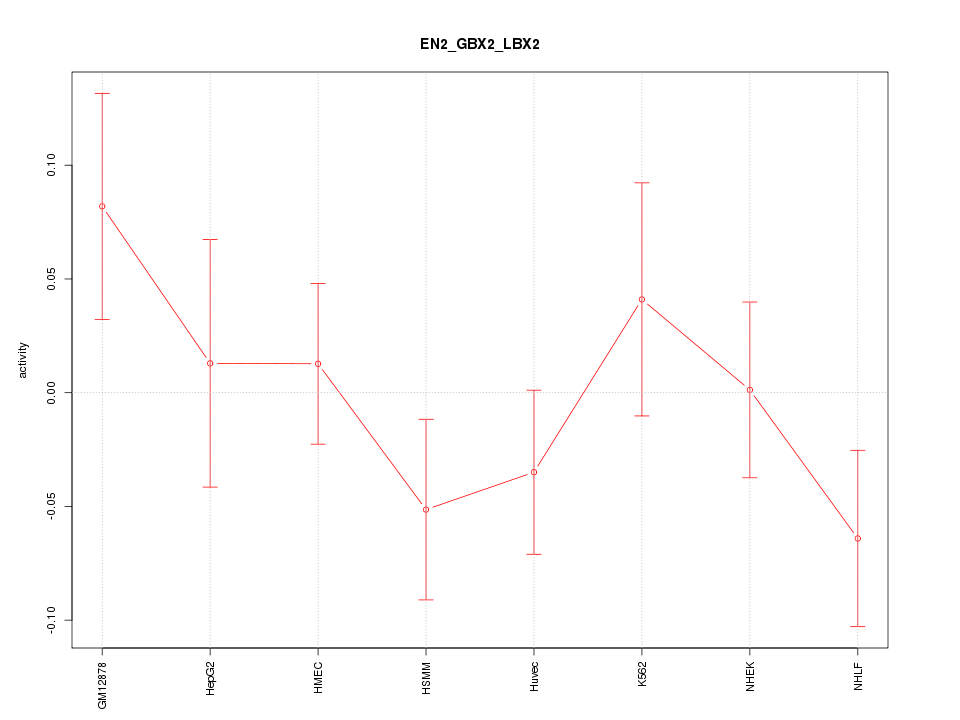
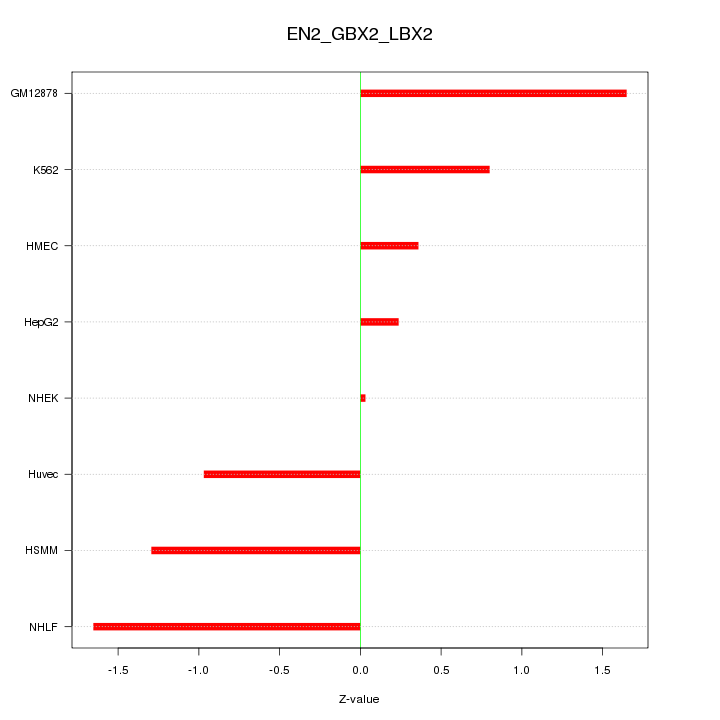
Motif ID: EN2_GBX2_LBX2
Z-value: 1.054
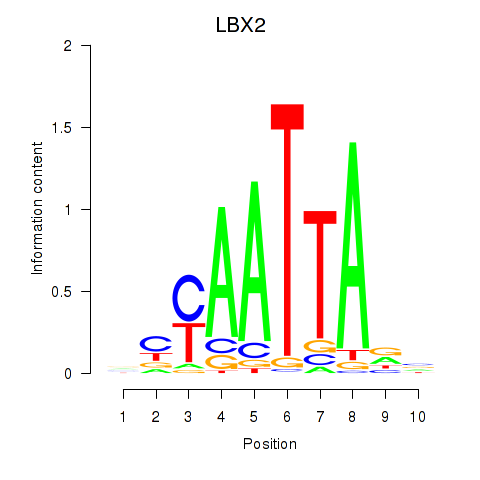
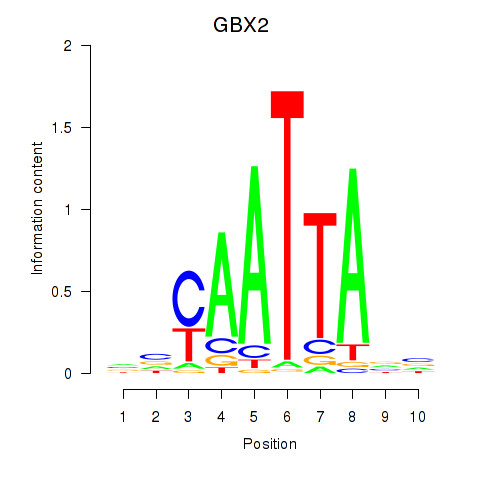
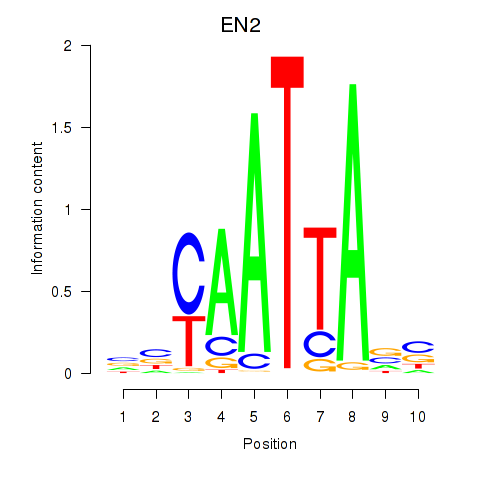
Transcription factors associated with EN2_GBX2_LBX2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| EN2 | ENSG00000164778.4 | EN2 |
| GBX2 | ENSG00000168505.6 | GBX2 |
| LBX2 | ENSG00000179528.11 | LBX2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.6 | GO:0043316 | natural killer cell activation involved in immune response(GO:0002323) cytotoxic T cell degranulation(GO:0043316) natural killer cell degranulation(GO:0043320) |
| 0.4 | 1.1 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.2 | 1.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.2 | 0.6 | GO:0060649 | nipple development(GO:0060618) mammary gland bud elongation(GO:0060649) nipple morphogenesis(GO:0060658) nipple sheath formation(GO:0060659) |
| 0.2 | 1.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.3 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 0.2 | GO:0045750 | obsolete positive regulation of S phase of mitotic cell cycle(GO:0045750) |
| 0.1 | 0.3 | GO:0032328 | alanine transport(GO:0032328) |
| 0.1 | 0.4 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) negative regulation of meiotic nuclear division(GO:0045835) regulation of meiosis I(GO:0060631) |
| 0.1 | 0.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) |
| 0.1 | 1.0 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.6 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.6 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.1 | 0.2 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 | 0.2 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.2 | GO:1904037 | positive regulation of epithelial cell apoptotic process(GO:1904037) positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.3 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.2 | GO:0060746 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 | 0.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.4 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.8 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.3 | GO:0060998 | regulation of dendritic spine development(GO:0060998) |
| 0.0 | 2.0 | GO:0031497 | chromatin assembly(GO:0031497) |
| 0.0 | 0.1 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.0 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 0.0 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.1 | GO:0032099 | negative regulation of appetite(GO:0032099) |
| 0.0 | 0.2 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.9 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.3 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.2 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.6 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.4 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.1 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) synaptic cleft(GO:0043083) |
| 0.1 | 0.9 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.1 | 4.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.7 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.0 | GO:0016363 | nuclear matrix(GO:0016363) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 0.9 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.4 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.1 | 1.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.5 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.3 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.6 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.2 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.1 | 0.2 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.4 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 1.2 | GO:0004221 | obsolete ubiquitin thiolesterase activity(GO:0004221) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 1.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.2 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.9 | GO:0016566 | obsolete specific transcriptional repressor activity(GO:0016566) |
| 0.0 | 0.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.0 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |