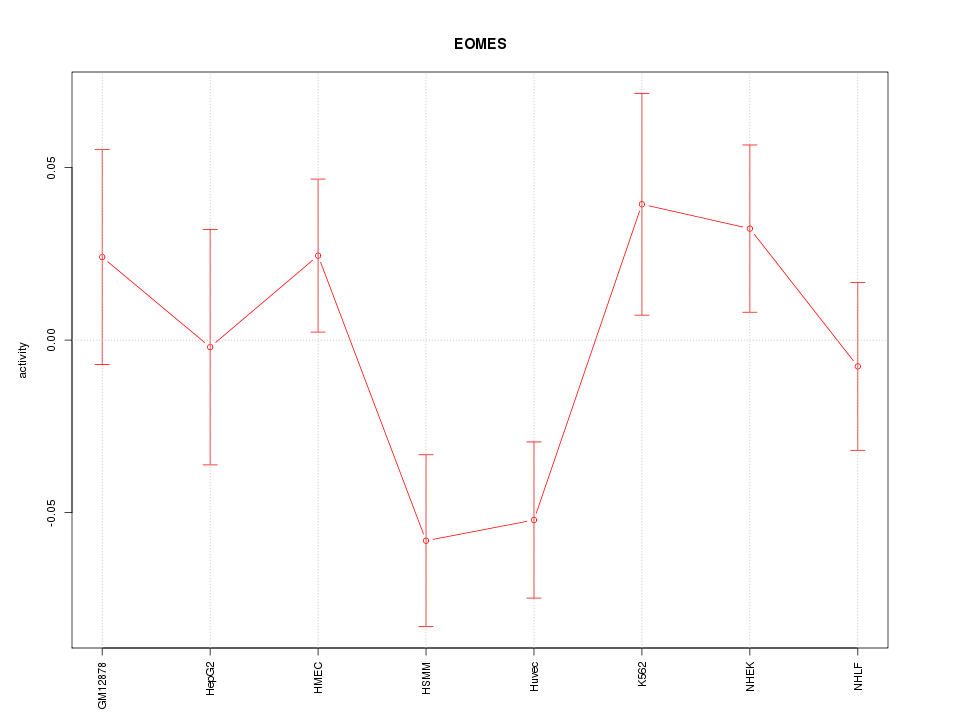
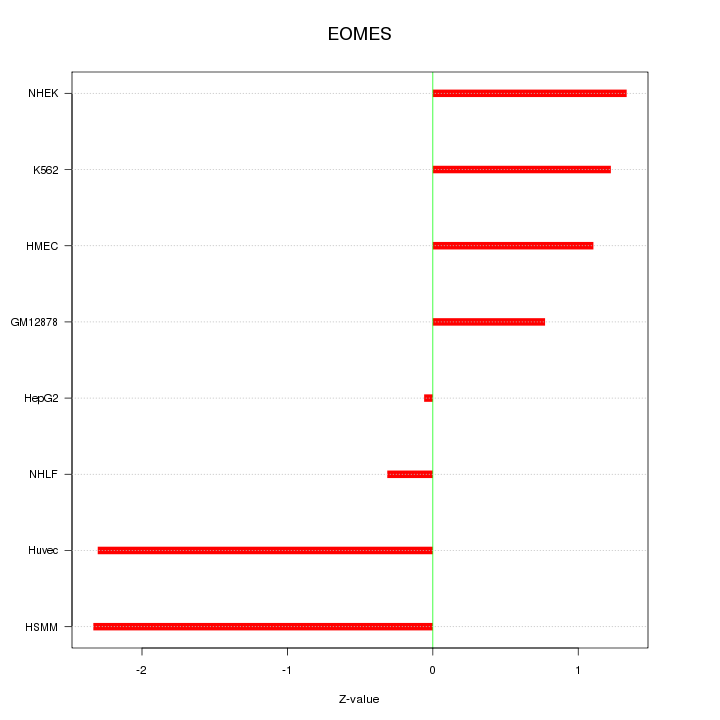
Motif ID: EOMES
Z-value: 1.412
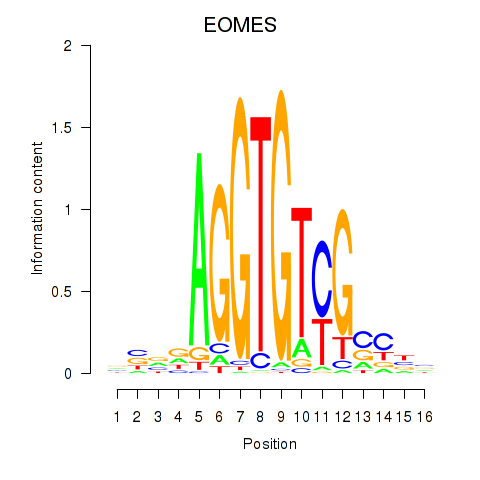
Transcription factors associated with EOMES:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| EOMES | ENSG00000163508.8 | EOMES |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 | 1.2 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.3 | 0.8 | GO:0071603 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) endothelial cell-cell adhesion(GO:0071603) |
| 0.2 | 0.7 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.2 | 0.6 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.2 | 0.6 | GO:1904742 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) regulation of telomeric DNA binding(GO:1904742) positive regulation of telomeric DNA binding(GO:1904744) |
| 0.2 | 1.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 0.6 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.2 | 0.5 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.2 | 1.0 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.2 | 0.5 | GO:0046080 | dUTP metabolic process(GO:0046080) |
| 0.2 | 0.5 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.1 | 0.4 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.1 | 0.2 | GO:0046060 | dATP metabolic process(GO:0046060) |
| 0.1 | 0.3 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 0.3 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 0.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.3 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.1 | 0.4 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 0.3 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 | 0.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.2 | GO:0045616 | regulation of keratinocyte differentiation(GO:0045616) |
| 0.1 | 0.3 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.1 | 0.4 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.7 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 0.3 | GO:0035283 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.3 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 | 1.8 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.1 | 0.3 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 0.7 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 0.3 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.2 | GO:0072053 | trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.1 | 0.2 | GO:0036260 | 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.1 | 0.4 | GO:2000189 | regulation of cholesterol homeostasis(GO:2000188) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.2 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 0.2 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.2 | GO:0015854 | guanine transport(GO:0015854) hypoxanthine transport(GO:0035344) thymine transport(GO:0035364) |
| 0.1 | 0.2 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.1 | 0.2 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 0.2 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.9 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.4 | GO:0060323 | head morphogenesis(GO:0060323) |
| 0.0 | 0.3 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.2 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.4 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.8 | GO:0045885 | obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.0 | 0.0 | GO:0048369 | lateral mesoderm development(GO:0048368) lateral mesoderm morphogenesis(GO:0048369) lateral mesoderm formation(GO:0048370) lateral mesodermal cell differentiation(GO:0048371) |
| 0.0 | 0.0 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.2 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.3 | GO:0046349 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.6 | GO:0050812 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 1.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0045759 | negative regulation of fatty acid beta-oxidation(GO:0031999) negative regulation of dopamine secretion(GO:0033602) negative regulation of action potential(GO:0045759) |
| 0.0 | 2.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.2 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.1 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.6 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 0.2 | GO:0045060 | negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.1 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.4 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.3 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.3 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.9 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.7 | GO:0000080 | mitotic G1 phase(GO:0000080) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.3 | GO:0001660 | fever generation(GO:0001660) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0072106 | regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.0 | 0.2 | GO:0045408 | regulation of interleukin-6 biosynthetic process(GO:0045408) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.5 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) |
| 0.0 | 2.6 | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.3 | GO:0002920 | regulation of humoral immune response(GO:0002920) |
| 0.0 | 0.1 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.1 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 3.0 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 0.2 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.3 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.2 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.3 | GO:0040023 | establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.3 | GO:0006541 | glutamine metabolic process(GO:0006541) |
| 0.0 | 0.6 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.2 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.0 | 0.9 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.0 | GO:1901642 | purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.7 | GO:0034976 | response to endoplasmic reticulum stress(GO:0034976) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.4 | GO:0050684 | regulation of mRNA processing(GO:0050684) regulation of mRNA metabolic process(GO:1903311) |
| 0.0 | 0.3 | GO:0001525 | angiogenesis(GO:0001525) |
| 0.0 | 0.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) INO80-type complex(GO:0097346) |
| 0.2 | 0.7 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.2 | 0.2 | GO:0019867 | outer membrane(GO:0019867) |
| 0.1 | 0.8 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.3 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 1.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.5 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 1.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.8 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.6 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.6 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.7 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) |
| 0.0 | 0.2 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0042405 | Mre11 complex(GO:0030870) nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.9 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.6 | GO:0042645 | mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:0016235 | aggresome(GO:0016235) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) exoribonuclease II activity(GO:0008859) |
| 0.3 | 0.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 0.7 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.2 | 0.7 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.2 | 0.7 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.2 | 0.6 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.2 | 0.5 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.1 | 0.9 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.5 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 1.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.3 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.8 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) alpha-catenin binding(GO:0045294) |
| 0.1 | 0.3 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.1 | 0.3 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.2 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.1 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.2 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.1 | 1.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.9 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 0.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 1.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.2 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.1 | 0.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.3 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 1.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.2 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.2 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.0 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.1 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.9 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 1.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 1.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.3 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.3 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) |
| 0.0 | 0.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0008753 | NADPH dehydrogenase (quinone) activity(GO:0008753) |
| 0.0 | 0.7 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.2 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 1.8 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 2.9 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.1 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 0.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 2.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) estrogen response element binding(GO:0034056) |
| 0.0 | 1.0 | GO:0031072 | heat shock protein binding(GO:0031072) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | ST_JAK_STAT_PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.0 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.0 | 0.4 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.8 | SIG_IL4RECEPTOR_IN_B_LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.6 | ST_G_ALPHA_I_PATHWAY | G alpha i Pathway |
| 0.0 | 0.3 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |