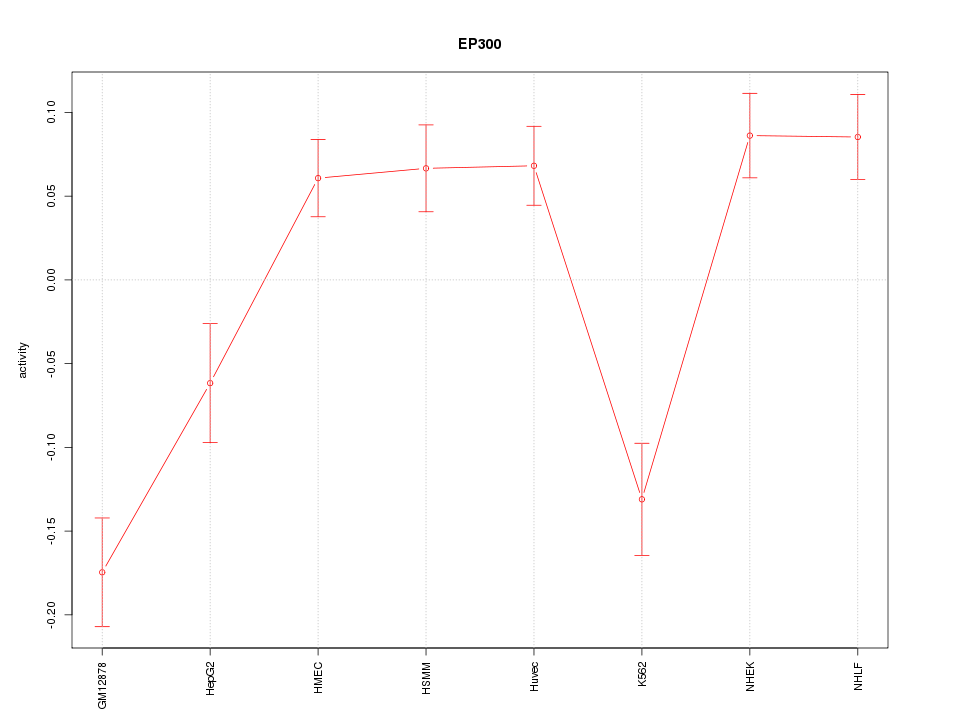
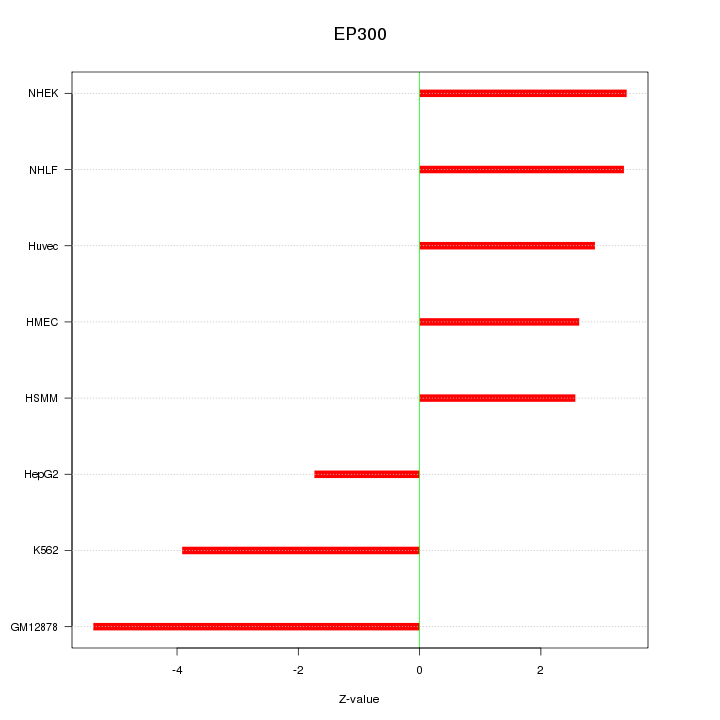
Motif ID: EP300
Z-value: 3.395
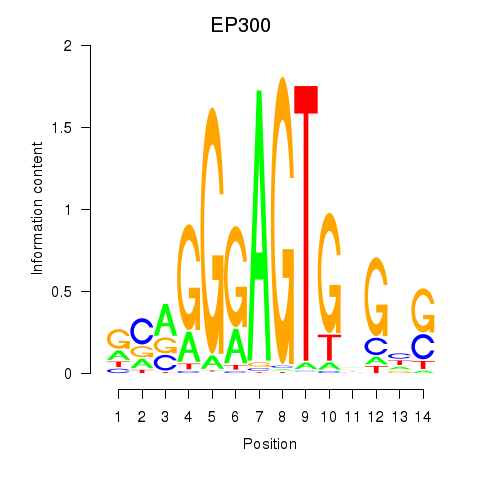
Transcription factors associated with EP300:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| EP300 | ENSG00000100393.9 | EP300 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 13.0 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 4.1 | 16.5 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 1.1 | 3.3 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 1.1 | 3.2 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 1.0 | 4.8 | GO:0015827 | tryptophan transport(GO:0015827) leucine import(GO:0060356) |
| 0.8 | 3.1 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.8 | 3.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.7 | 2.8 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 0.7 | 9.8 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.6 | 2.6 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) long-term synaptic potentiation(GO:0060291) |
| 0.6 | 1.7 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.6 | 1.7 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) negative regulation of protein glycosylation(GO:0060051) |
| 0.5 | 4.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.5 | 4.1 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.4 | 1.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.4 | 1.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.4 | 1.1 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.3 | 2.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.3 | 1.0 | GO:0046604 | regulation of mitotic centrosome separation(GO:0046602) positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.3 | 16.3 | GO:0007045 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.3 | 0.9 | GO:0032938 | negative regulation of translation in response to oxidative stress(GO:0032938) regulation of translation in response to oxidative stress(GO:0043556) |
| 0.3 | 0.9 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.3 | 1.7 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.3 | 4.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.3 | 1.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.3 | 1.1 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.3 | 5.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.2 | 3.9 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.2 | 1.0 | GO:0046070 | dGTP metabolic process(GO:0046070) purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.2 | 2.1 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.2 | 1.0 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 1.6 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.2 | 0.9 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.2 | 0.6 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.2 | 4.1 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.2 | 0.8 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.2 | 0.9 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.1 | 0.7 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 5.3 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.1 | 0.4 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 2.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 4.9 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 1.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 1.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 | 5.6 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.1 | 1.0 | GO:0006959 | humoral immune response(GO:0006959) |
| 0.1 | 0.3 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 0.5 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 1.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 3.2 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.1 | 0.5 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 0.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 1.0 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.1 | 0.3 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.1 | 1.4 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 1.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.5 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.1 | 0.5 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.1 | 1.0 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 6.3 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.4 | GO:0044349 | nucleotide-excision repair, DNA damage removal(GO:0000718) DNA excision(GO:0044349) |
| 0.1 | 0.1 | GO:0006490 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 0.8 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 0.5 | GO:0051924 | regulation of calcium ion transport(GO:0051924) |
| 0.1 | 1.2 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 2.6 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 7.4 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.6 | GO:0009790 | embryo development(GO:0009790) |
| 0.0 | 7.7 | GO:0034329 | cell junction assembly(GO:0034329) |
| 0.0 | 1.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.6 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.3 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.8 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.3 | GO:0016559 | peroxisome membrane biogenesis(GO:0016557) peroxisome fission(GO:0016559) |
| 0.0 | 0.6 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 1.5 | GO:0042384 | cilium assembly(GO:0042384) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 2.6 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.2 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.0 | 0.3 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.3 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.7 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 0.8 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.8 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.6 | GO:0031529 | lamellipodium assembly(GO:0030032) ruffle organization(GO:0031529) |
| 0.0 | 5.5 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.3 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.4 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 4.1 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.6 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.2 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.0 | 0.5 | GO:0007389 | pattern specification process(GO:0007389) |
| 0.0 | 4.5 | GO:0007015 | actin filament organization(GO:0007015) |
| 0.0 | 0.7 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 1.7 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 1.8 | GO:0043473 | pigmentation(GO:0043473) |
| 0.0 | 1.5 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) actin-mediated cell contraction(GO:0070252) |
| 0.0 | 1.2 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.5 | GO:0045446 | endothelial cell differentiation(GO:0045446) |
| 0.0 | 4.7 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.7 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.4 | GO:0031055 | DNA replication-independent nucleosome assembly(GO:0006336) chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) DNA replication-independent nucleosome organization(GO:0034724) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 3.4 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 2.7 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 0.1 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 | 0.1 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.8 | GO:0007163 | establishment or maintenance of cell polarity(GO:0007163) |
| 0.0 | 0.0 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 | 0.4 | GO:0042044 | water transport(GO:0006833) fluid transport(GO:0042044) |
| 0.0 | 0.2 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.3 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.7 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 0.2 | GO:0051181 | cofactor transport(GO:0051181) |
| 0.0 | 0.2 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.0 | GO:0045060 | negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.1 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 2.0 | GO:0006887 | exocytosis(GO:0006887) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 19.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.9 | 16.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.6 | 2.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.6 | 12.6 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.6 | 1.7 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.3 | 3.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.3 | 3.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.3 | 0.9 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.3 | 1.5 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 1.0 | GO:0043219 | lateral loop(GO:0043219) |
| 0.2 | 14.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 13.4 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.2 | 0.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.2 | 4.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 1.7 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.1 | 2.4 | GO:0001527 | microfibril(GO:0001527) |
| 0.1 | 1.0 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 2.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 3.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.4 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.1 | 1.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.6 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 1.2 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.1 | 1.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 8.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 2.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 5.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 5.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 2.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 3.3 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 4.1 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 3.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.9 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 2.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.2 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.9 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 1.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.6 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 1.0 | GO:0019717 | obsolete synaptosome(GO:0019717) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 1.0 | 3.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.8 | 3.3 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) |
| 0.8 | 16.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.7 | 2.8 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.6 | 2.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.6 | 3.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.5 | 4.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.5 | 1.4 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.4 | 2.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.4 | 2.9 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.3 | 1.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.3 | 1.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.3 | 3.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.3 | 4.1 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.3 | 1.4 | GO:0080030 | methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.3 | 1.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.3 | 0.8 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.2 | 1.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.2 | 0.9 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 3.9 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.2 | 1.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 3.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.2 | 1.6 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.2 | 0.7 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.2 | 2.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 1.0 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 1.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 0.8 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.2 | 16.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 0.5 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.2 | 1.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 3.0 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.1 | 0.9 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 1.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 2.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.9 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 2.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 1.7 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.8 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.1 | 1.6 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 1.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.6 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 8.2 | GO:0051287 | NAD binding(GO:0051287) |
| 0.1 | 1.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 1.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.3 | GO:0017108 | crossover junction endodeoxyribonuclease activity(GO:0008821) 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 1.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 1.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.6 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 2.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 1.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 4.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.4 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 3.1 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 0.3 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 1.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.5 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 2.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 14.9 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 0.5 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 1.6 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 1.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 7.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.6 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.8 | GO:0001104 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.3 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 2.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.4 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 5.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 5.2 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 2.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 9.0 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 5.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.0 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0051184 | cofactor transporter activity(GO:0051184) |
| 0.0 | 0.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 2.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.1 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.9 | SIG_CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 16.9 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |
| 0.1 | 1.2 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.7 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | SIG_IL4RECEPTOR_IN_B_LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |