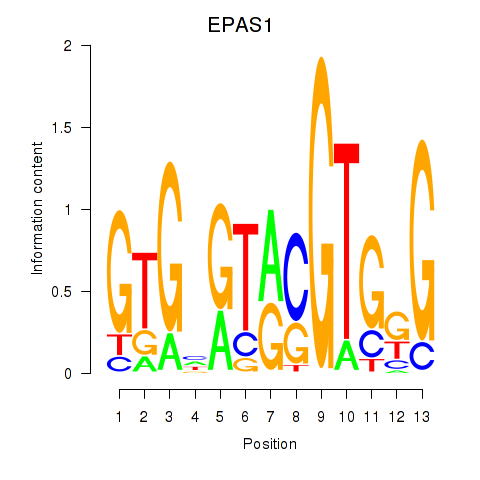
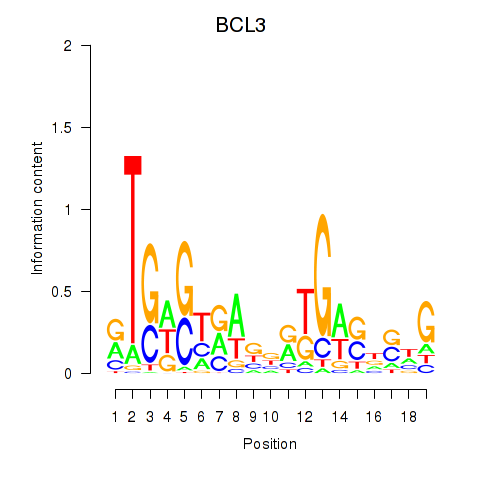
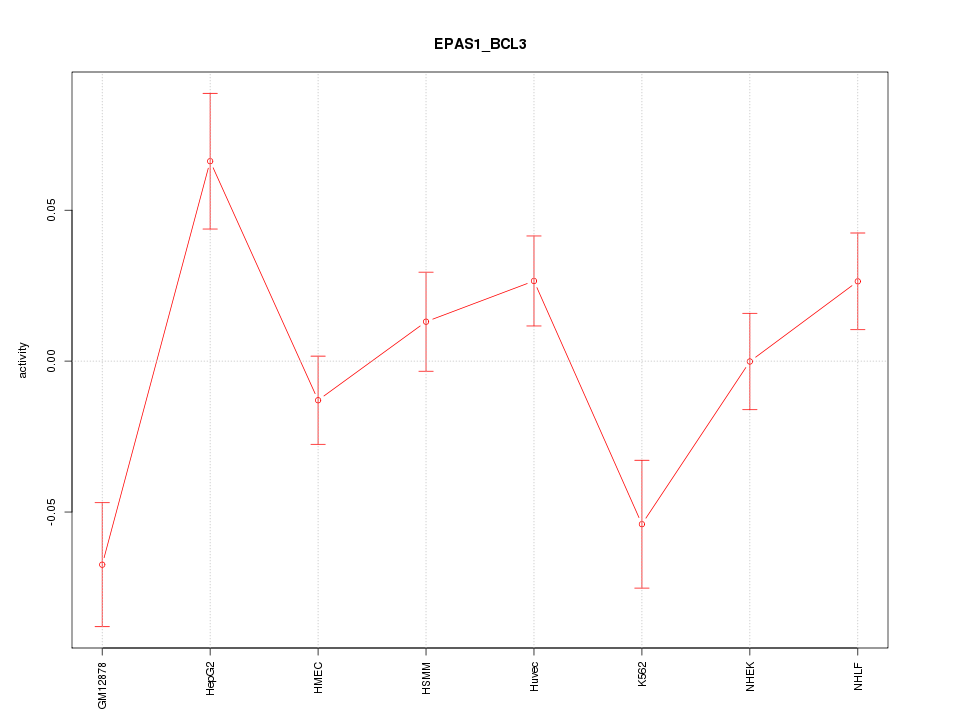
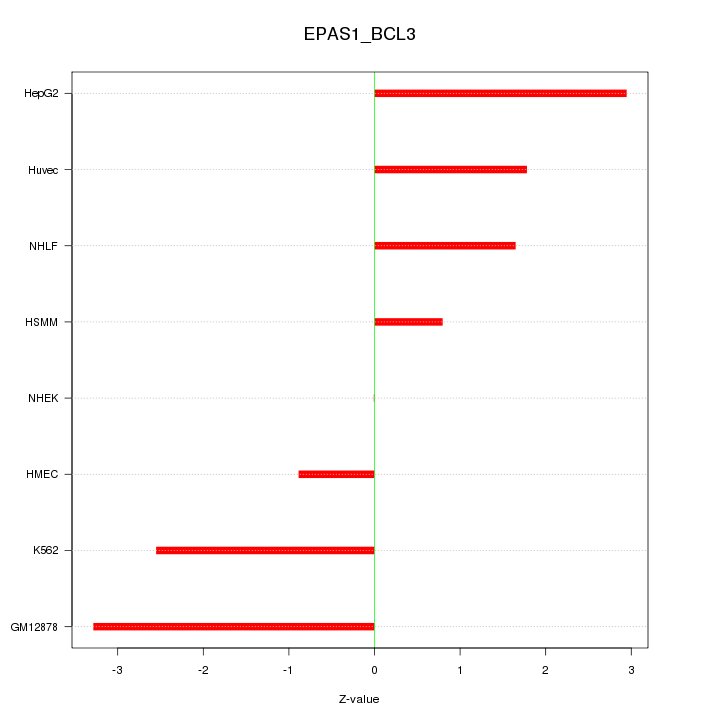
| 1.7 |
5.1 |
GO:0010902 |
positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) positive regulation of phospholipid catabolic process(GO:0060697) |
| 1.3 |
7.5 |
GO:0046882 |
negative regulation of gonadotropin secretion(GO:0032277) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.9 |
3.7 |
GO:0045048 |
protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.9 |
2.8 |
GO:0002514 |
B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.7 |
3.6 |
GO:0006927 |
obsolete transformed cell apoptotic process(GO:0006927) |
| 0.7 |
4.1 |
GO:0060710 |
chorio-allantoic fusion(GO:0060710) |
| 0.7 |
2.0 |
GO:0010757 |
chronological cell aging(GO:0001300) negative regulation of plasminogen activation(GO:0010757) negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.7 |
3.3 |
GO:1901031 |
regulation of cellular response to oxidative stress(GO:1900407) regulation of response to reactive oxygen species(GO:1901031) regulation of response to oxidative stress(GO:1902882) |
| 0.6 |
1.9 |
GO:0001907 |
cytolysis by symbiont of host cells(GO:0001897) killing by symbiont of host cells(GO:0001907) hemolysis by symbiont of host erythrocytes(GO:0019836) disruption by symbiont of host cell(GO:0044004) hemolysis in other organism(GO:0044179) positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) cytolysis in other organism(GO:0051715) cytolysis in other organism involved in symbiotic interaction(GO:0051801) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.6 |
7.0 |
GO:0060762 |
regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.5 |
2.2 |
GO:0034442 |
plasma lipoprotein particle oxidation(GO:0034441) regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) lipoprotein modification(GO:0042160) lipoprotein oxidation(GO:0042161) |
| 0.5 |
1.6 |
GO:0002374 |
cytokine secretion involved in immune response(GO:0002374) regulation of cytokine secretion involved in immune response(GO:0002739) negative regulation of cytokine secretion involved in immune response(GO:0002740) regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 0.5 |
1.6 |
GO:0002887 |
wound healing involved in inflammatory response(GO:0002246) negative regulation of myeloid leukocyte mediated immunity(GO:0002887) heme oxidation(GO:0006788) negative regulation of leukocyte degranulation(GO:0043301) inflammatory response to wounding(GO:0090594) |
| 0.5 |
1.5 |
GO:0048389 |
intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) bud dilation involved in lung branching(GO:0060503) branch elongation involved in ureteric bud branching(GO:0060681) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) pulmonary artery morphogenesis(GO:0061156) cell proliferation involved in mesonephros development(GO:0061209) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) negative regulation of mesonephros development(GO:0061218) pattern specification involved in mesonephros development(GO:0061227) renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) BMP signaling pathway involved in nephric duct formation(GO:0071893) glomerular visceral epithelial cell development(GO:0072015) comma-shaped body morphogenesis(GO:0072049) regulation of branch elongation involved in ureteric bud branching(GO:0072095) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) glomerulus morphogenesis(GO:0072102) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) nephric duct formation(GO:0072179) ureter urothelium development(GO:0072190) ureter epithelial cell differentiation(GO:0072192) ureter morphogenesis(GO:0072197) metanephric comma-shaped body morphogenesis(GO:0072278) glomerular epithelial cell development(GO:0072310) negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.5 |
1.5 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.4 |
1.3 |
GO:1990170 |
detoxification of cadmium ion(GO:0071585) stress response to cadmium ion(GO:1990170) |
| 0.4 |
1.9 |
GO:0007614 |
short-term memory(GO:0007614) |
| 0.3 |
2.0 |
GO:0032460 |
negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.3 |
1.6 |
GO:0060662 |
tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.3 |
0.9 |
GO:1904729 |
regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.3 |
1.2 |
GO:0030037 |
actin filament reorganization involved in cell cycle(GO:0030037) actin filament reorganization(GO:0090527) |
| 0.3 |
1.1 |
GO:0034959 |
neuropeptide catabolic process(GO:0010813) substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.3 |
2.6 |
GO:0055003 |
cardiac myofibril assembly(GO:0055003) |
| 0.3 |
3.6 |
GO:0001945 |
lymph vessel development(GO:0001945) |
| 0.3 |
3.3 |
GO:1904893 |
negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 |
0.7 |
GO:0070672 |
response to interleukin-15(GO:0070672) |
| 0.2 |
3.7 |
GO:0034446 |
substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.2 |
3.2 |
GO:0032331 |
negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.2 |
0.7 |
GO:0048319 |
axial mesoderm morphogenesis(GO:0048319) |
| 0.2 |
0.7 |
GO:0048861 |
leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.2 |
1.1 |
GO:0010890 |
positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.2 |
0.4 |
GO:0060394 |
negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 |
0.8 |
GO:0031340 |
positive regulation of vesicle fusion(GO:0031340) |
| 0.2 |
0.2 |
GO:0060677 |
ureteric bud elongation(GO:0060677) |
| 0.2 |
0.4 |
GO:0002890 |
negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.2 |
0.3 |
GO:0060214 |
endocardium formation(GO:0060214) |
| 0.1 |
1.3 |
GO:0006546 |
glycine catabolic process(GO:0006546) |
| 0.1 |
0.8 |
GO:0006562 |
proline catabolic process(GO:0006562) |
| 0.1 |
1.5 |
GO:0046628 |
positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 |
0.4 |
GO:0045618 |
positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.1 |
0.4 |
GO:0009635 |
response to herbicide(GO:0009635) |
| 0.1 |
0.4 |
GO:0002575 |
basophil chemotaxis(GO:0002575) |
| 0.1 |
0.4 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |
| 0.1 |
0.6 |
GO:0070245 |
positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 |
0.3 |
GO:0050758 |
thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 |
0.7 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.1 |
0.4 |
GO:0072385 |
minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 |
0.8 |
GO:0071731 |
response to nitric oxide(GO:0071731) |
| 0.1 |
0.3 |
GO:0008298 |
intracellular mRNA localization(GO:0008298) |
| 0.1 |
0.4 |
GO:0009109 |
coenzyme catabolic process(GO:0009109) |
| 0.1 |
8.0 |
GO:0031668 |
cellular response to extracellular stimulus(GO:0031668) |
| 0.1 |
0.5 |
GO:0060164 |
regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 |
1.4 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.1 |
0.7 |
GO:0035058 |
nonmotile primary cilium assembly(GO:0035058) |
| 0.1 |
1.9 |
GO:0032012 |
regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 |
1.8 |
GO:0003407 |
neural retina development(GO:0003407) |
| 0.1 |
3.9 |
GO:0030520 |
intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.1 |
0.7 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 |
1.6 |
GO:0043586 |
tongue development(GO:0043586) |
| 0.1 |
0.2 |
GO:0048520 |
positive regulation of behavior(GO:0048520) |
| 0.1 |
0.4 |
GO:0045945 |
positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.1 |
0.5 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.1 |
0.9 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.1 |
0.3 |
GO:0002158 |
osteoclast proliferation(GO:0002158) |
| 0.1 |
0.3 |
GO:0009153 |
purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 0.1 |
0.3 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 |
0.4 |
GO:0035444 |
nickel cation transport(GO:0015675) vanadium ion transport(GO:0015676) ferrous iron transport(GO:0015684) lead ion transport(GO:0015692) nickel cation transmembrane transport(GO:0035444) ferrous iron import(GO:0070627) iron ion import(GO:0097286) |
| 0.1 |
0.4 |
GO:0009629 |
response to gravity(GO:0009629) |
| 0.1 |
0.2 |
GO:0035120 |
post-embryonic appendage morphogenesis(GO:0035120) |
| 0.1 |
5.6 |
GO:0006821 |
chloride transport(GO:0006821) |
| 0.1 |
0.2 |
GO:0018874 |
benzoate metabolic process(GO:0018874) |
| 0.1 |
0.1 |
GO:0090206 |
negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 |
0.4 |
GO:0002220 |
innate immune response activating cell surface receptor signaling pathway(GO:0002220) |
| 0.1 |
1.4 |
GO:0009086 |
methionine biosynthetic process(GO:0009086) |
| 0.1 |
0.3 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.1 |
0.2 |
GO:0045760 |
positive regulation of action potential(GO:0045760) |
| 0.1 |
0.4 |
GO:0009450 |
gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.1 |
0.5 |
GO:0035313 |
wound healing, spreading of epidermal cells(GO:0035313) wound healing, spreading of cells(GO:0044319) epiboly(GO:0090504) epiboly involved in wound healing(GO:0090505) |
| 0.1 |
1.0 |
GO:0006750 |
glutathione biosynthetic process(GO:0006750) |
| 0.1 |
2.1 |
GO:0007566 |
embryo implantation(GO:0007566) |
| 0.1 |
2.1 |
GO:0046847 |
filopodium assembly(GO:0046847) |
| 0.1 |
0.1 |
GO:0007403 |
glial cell fate determination(GO:0007403) |
| 0.1 |
0.3 |
GO:0002052 |
positive regulation of neuroblast proliferation(GO:0002052) |
| 0.1 |
0.9 |
GO:0008635 |
activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 |
0.4 |
GO:0030277 |
epithelial structure maintenance(GO:0010669) maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 |
0.6 |
GO:0019885 |
antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 |
1.9 |
GO:0015914 |
phospholipid transport(GO:0015914) |
| 0.1 |
5.6 |
GO:0007565 |
female pregnancy(GO:0007565) |
| 0.1 |
2.3 |
GO:0045576 |
mast cell activation(GO:0045576) |
| 0.1 |
1.7 |
GO:0030866 |
cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 |
0.1 |
GO:0010727 |
negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 |
2.7 |
GO:0032526 |
response to retinoic acid(GO:0032526) |
| 0.0 |
2.7 |
GO:0055088 |
lipid homeostasis(GO:0055088) |
| 0.0 |
0.5 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 |
0.1 |
GO:0050917 |
sensory perception of umami taste(GO:0050917) |
| 0.0 |
0.2 |
GO:0032465 |
regulation of cytokinesis(GO:0032465) |
| 0.0 |
2.8 |
GO:0007569 |
cell aging(GO:0007569) |
| 0.0 |
0.3 |
GO:0090205 |
positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 |
0.2 |
GO:0050968 |
detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 |
0.1 |
GO:0072334 |
UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) nucleotide transmembrane transport(GO:1901679) |
| 0.0 |
0.4 |
GO:0051016 |
barbed-end actin filament capping(GO:0051016) |
| 0.0 |
0.1 |
GO:0021670 |
lateral ventricle development(GO:0021670) |
| 0.0 |
0.8 |
GO:0009812 |
flavonoid metabolic process(GO:0009812) |
| 0.0 |
0.3 |
GO:0042491 |
auditory receptor cell differentiation(GO:0042491) |
| 0.0 |
1.8 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
0.3 |
GO:0097384 |
cellular lipid biosynthetic process(GO:0097384) |
| 0.0 |
0.2 |
GO:0035246 |
peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 |
0.1 |
GO:0048702 |
embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 |
0.1 |
GO:0050653 |
chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 |
0.2 |
GO:0008653 |
lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 |
0.1 |
GO:0001880 |
Mullerian duct regression(GO:0001880) |
| 0.0 |
0.1 |
GO:0033687 |
osteoblast proliferation(GO:0033687) |
| 0.0 |
0.1 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.0 |
0.2 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.1 |
GO:0006540 |
glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 |
0.7 |
GO:0046470 |
phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 |
0.1 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.1 |
GO:0014912 |
negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 |
1.0 |
GO:0007585 |
respiratory gaseous exchange(GO:0007585) |
| 0.0 |
0.1 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.0 |
0.2 |
GO:0007423 |
sensory organ development(GO:0007423) |
| 0.0 |
1.2 |
GO:0009791 |
post-embryonic development(GO:0009791) |
| 0.0 |
3.7 |
GO:0006869 |
lipid transport(GO:0006869) |
| 0.0 |
0.1 |
GO:0030147 |
obsolete natriuresis(GO:0030147) |
| 0.0 |
0.8 |
GO:0007189 |
adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 |
0.6 |
GO:0006921 |
cellular component disassembly involved in execution phase of apoptosis(GO:0006921) execution phase of apoptosis(GO:0097194) |
| 0.0 |
0.0 |
GO:0032328 |
alanine transport(GO:0032328) |
| 0.0 |
0.1 |
GO:0070309 |
lens fiber cell morphogenesis(GO:0070309) |
| 0.0 |
0.1 |
GO:0006295 |
nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 |
0.2 |
GO:0045070 |
positive regulation of viral genome replication(GO:0045070) |
| 0.0 |
1.4 |
GO:0007229 |
integrin-mediated signaling pathway(GO:0007229) |
| 0.0 |
0.3 |
GO:0042994 |
cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 |
0.1 |
GO:0001941 |
postsynaptic membrane organization(GO:0001941) |
| 0.0 |
0.4 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 |
0.1 |
GO:0006069 |
ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 |
0.2 |
GO:0019228 |
neuronal action potential(GO:0019228) |
| 0.0 |
0.4 |
GO:0008154 |
actin polymerization or depolymerization(GO:0008154) |
| 0.0 |
0.1 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.4 |
GO:0090630 |
activation of GTPase activity(GO:0090630) |
| 0.0 |
0.2 |
GO:0043113 |
receptor clustering(GO:0043113) |
| 0.0 |
0.1 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.1 |
GO:0045163 |
clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 |
0.1 |
GO:0046642 |
negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 |
0.1 |
GO:0070120 |
ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 |
0.3 |
GO:0007263 |
nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 |
1.0 |
GO:0008064 |
regulation of actin polymerization or depolymerization(GO:0008064) |
| 0.0 |
0.4 |
GO:0051789 |
obsolete response to protein(GO:0051789) |
| 0.0 |
0.2 |
GO:0007044 |
cell-substrate junction assembly(GO:0007044) |
| 0.0 |
0.0 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 |
0.1 |
GO:0001539 |
cilium or flagellum-dependent cell motility(GO:0001539) |
| 0.0 |
0.0 |
GO:0045601 |
regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 |
0.4 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
0.3 |
GO:0015949 |
nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 |
0.2 |
GO:0006783 |
heme biosynthetic process(GO:0006783) |
| 0.0 |
0.3 |
GO:0005980 |
glycogen catabolic process(GO:0005980) |
| 0.0 |
0.0 |
GO:0043030 |
regulation of macrophage activation(GO:0043030) |
| 0.0 |
0.0 |
GO:0001507 |
acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 |
0.1 |
GO:0006198 |
cAMP catabolic process(GO:0006198) |