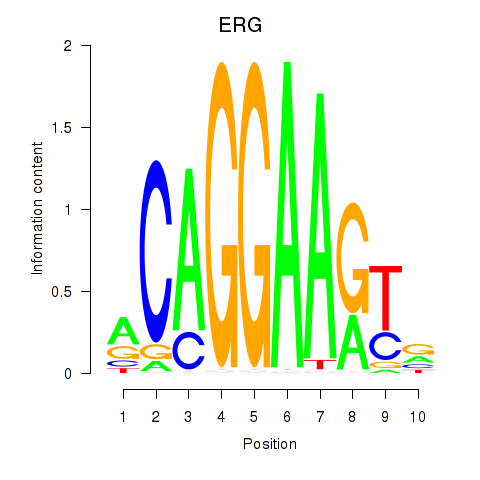
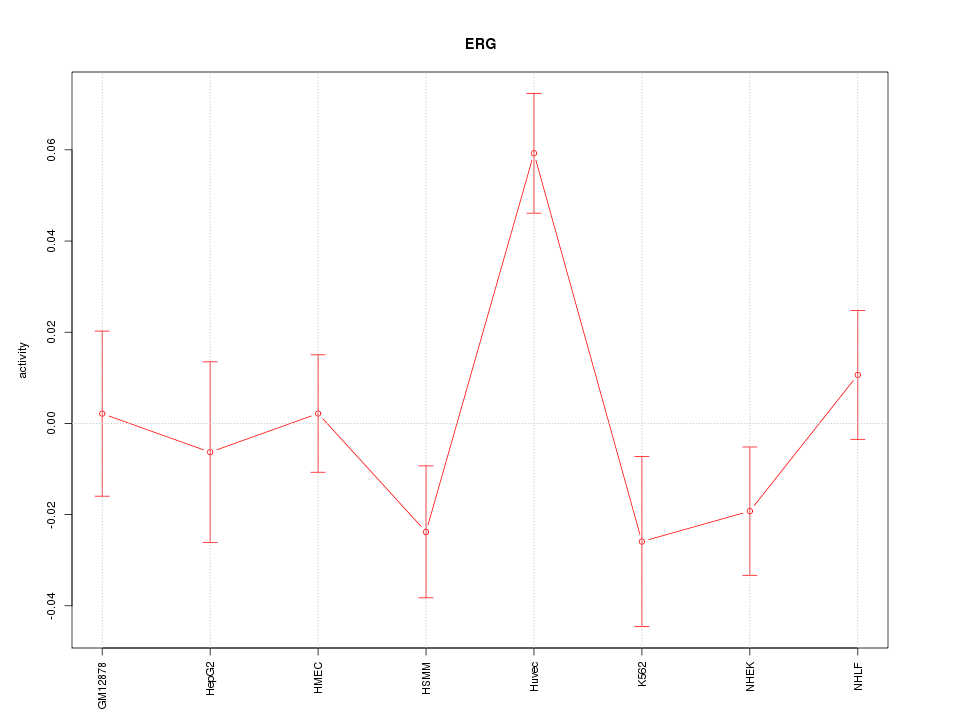
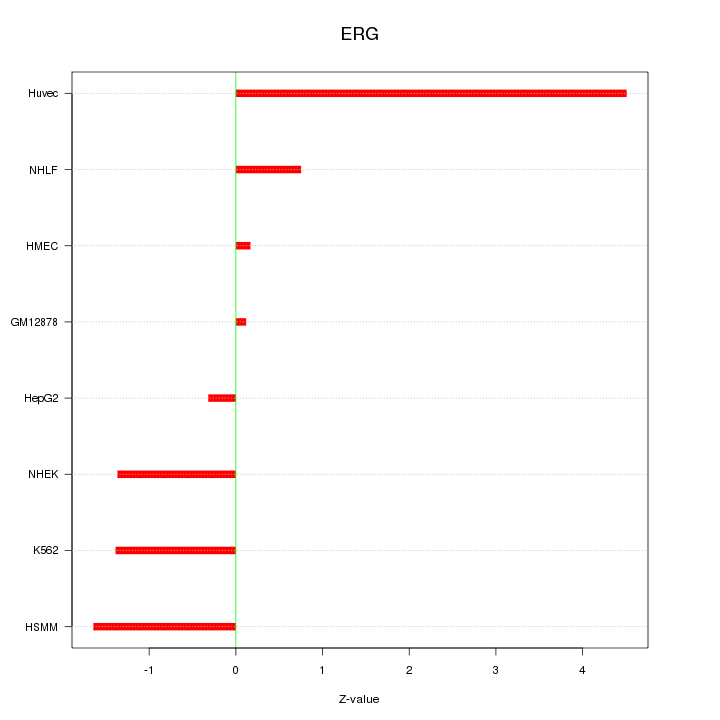
| 1.8 |
5.5 |
GO:0001300 |
chronological cell aging(GO:0001300) |
| 0.5 |
1.0 |
GO:0006154 |
adenosine catabolic process(GO:0006154) |
| 0.5 |
2.0 |
GO:0006663 |
platelet activating factor biosynthetic process(GO:0006663) positive regulation of vesicle fusion(GO:0031340) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.4 |
1.7 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.4 |
1.3 |
GO:0071380 |
cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.4 |
2.6 |
GO:0043615 |
astrocyte cell migration(GO:0043615) |
| 0.3 |
0.7 |
GO:0045715 |
negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.3 |
1.7 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.3 |
1.3 |
GO:0010566 |
regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.3 |
1.3 |
GO:0030913 |
paranodal junction assembly(GO:0030913) |
| 0.3 |
2.2 |
GO:0045079 |
negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.3 |
1.5 |
GO:0032497 |
detection of lipopolysaccharide(GO:0032497) |
| 0.3 |
0.3 |
GO:0002901 |
mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.3 |
1.1 |
GO:0001973 |
adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.3 |
2.0 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.3 |
0.6 |
GO:0010757 |
negative regulation of plasminogen activation(GO:0010757) |
| 0.3 |
1.6 |
GO:0060263 |
regulation of respiratory burst(GO:0060263) |
| 0.3 |
0.8 |
GO:0006051 |
mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.2 |
2.2 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.2 |
1.7 |
GO:0016559 |
peroxisome fission(GO:0016559) |
| 0.2 |
0.5 |
GO:0002043 |
blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.2 |
0.7 |
GO:0006427 |
histidyl-tRNA aminoacylation(GO:0006427) |
| 0.2 |
1.1 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 |
1.7 |
GO:0050872 |
white fat cell differentiation(GO:0050872) |
| 0.2 |
0.6 |
GO:0002523 |
leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.2 |
0.7 |
GO:0034959 |
neuropeptide catabolic process(GO:0010813) substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.2 |
0.2 |
GO:0019276 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.2 |
5.0 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.2 |
0.2 |
GO:0060632 |
regulation of microtubule-based movement(GO:0060632) |
| 0.2 |
5.0 |
GO:0030224 |
monocyte differentiation(GO:0030224) |
| 0.2 |
0.5 |
GO:0010836 |
negative regulation of protein ADP-ribosylation(GO:0010836) negative regulation of protein glycosylation(GO:0060051) |
| 0.2 |
2.0 |
GO:0006027 |
glycosaminoglycan catabolic process(GO:0006027) |
| 0.2 |
0.7 |
GO:0045048 |
protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.2 |
0.5 |
GO:0033278 |
cell proliferation in midbrain(GO:0033278) |
| 0.2 |
1.6 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.2 |
2.4 |
GO:0034446 |
substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.2 |
1.3 |
GO:0007185 |
transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.2 |
4.3 |
GO:0072378 |
blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 |
0.6 |
GO:1904037 |
positive regulation of epithelial cell apoptotic process(GO:1904037) positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.2 |
0.8 |
GO:0048680 |
positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.1 |
0.4 |
GO:0060178 |
exocyst assembly(GO:0001927) regulation of exocyst assembly(GO:0001928) exocyst localization(GO:0051601) regulation of exocyst localization(GO:0060178) |
| 0.1 |
0.4 |
GO:0046586 |
regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 |
0.9 |
GO:0032460 |
negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 |
0.4 |
GO:0030163 |
protein catabolic process(GO:0030163) |
| 0.1 |
0.1 |
GO:0051592 |
response to calcium ion(GO:0051592) |
| 0.1 |
0.8 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) positive regulation of adherens junction organization(GO:1903393) |
| 0.1 |
0.4 |
GO:0010982 |
regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 |
0.7 |
GO:0017055 |
negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.1 |
0.5 |
GO:0009956 |
radial pattern formation(GO:0009956) |
| 0.1 |
0.5 |
GO:0019046 |
release from viral latency(GO:0019046) |
| 0.1 |
0.1 |
GO:1901984 |
negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.1 |
0.4 |
GO:0019371 |
cyclooxygenase pathway(GO:0019371) |
| 0.1 |
0.6 |
GO:0032933 |
SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 |
0.5 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 |
0.7 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.1 |
1.9 |
GO:0060390 |
regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.1 |
0.5 |
GO:0009609 |
response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.1 |
0.6 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.1 |
0.3 |
GO:0060831 |
smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 |
0.7 |
GO:0051665 |
membrane raft localization(GO:0051665) |
| 0.1 |
0.5 |
GO:0061025 |
membrane fusion(GO:0061025) |
| 0.1 |
0.4 |
GO:0060037 |
pharyngeal system development(GO:0060037) |
| 0.1 |
0.2 |
GO:0050665 |
hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.1 |
1.0 |
GO:0046051 |
GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.1 |
1.6 |
GO:0000090 |
mitotic anaphase(GO:0000090) |
| 0.1 |
1.9 |
GO:0045026 |
plasma membrane fusion(GO:0045026) |
| 0.1 |
0.3 |
GO:0006922 |
obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.1 |
0.3 |
GO:0032074 |
negative regulation of nuclease activity(GO:0032074) |
| 0.1 |
0.7 |
GO:0032897 |
negative regulation of viral transcription(GO:0032897) |
| 0.1 |
1.3 |
GO:0050650 |
chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 |
0.4 |
GO:0002313 |
mature B cell differentiation involved in immune response(GO:0002313) |
| 0.1 |
0.2 |
GO:0006526 |
arginine biosynthetic process(GO:0006526) |
| 0.1 |
0.1 |
GO:0045618 |
positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.1 |
0.3 |
GO:0050703 |
interleukin-1 alpha secretion(GO:0050703) |
| 0.1 |
0.5 |
GO:0032007 |
negative regulation of TOR signaling(GO:0032007) |
| 0.1 |
0.5 |
GO:0006290 |
pyrimidine dimer repair(GO:0006290) |
| 0.1 |
0.3 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) cytoplasmic translation(GO:0002181) cytoplasmic translational initiation(GO:0002183) |
| 0.1 |
0.8 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.1 |
0.2 |
GO:0071502 |
cellular response to temperature stimulus(GO:0071502) |
| 0.1 |
1.3 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.1 |
0.2 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 |
0.9 |
GO:0032331 |
negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 |
2.2 |
GO:0042116 |
macrophage activation(GO:0042116) |
| 0.1 |
0.5 |
GO:0032472 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) Golgi calcium ion transport(GO:0032472) manganese ion homeostasis(GO:0055071) |
| 0.1 |
0.2 |
GO:0002291 |
T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 |
0.2 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.1 |
0.6 |
GO:0003065 |
regulation of heart rate by chemical signal(GO:0003062) positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 |
0.4 |
GO:0046398 |
UDP-glucuronate biosynthetic process(GO:0006065) UDP-glucuronate metabolic process(GO:0046398) |
| 0.1 |
1.2 |
GO:0051016 |
barbed-end actin filament capping(GO:0051016) |
| 0.1 |
0.4 |
GO:0046642 |
negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 |
1.9 |
GO:0043496 |
regulation of protein homodimerization activity(GO:0043496) |
| 0.1 |
1.5 |
GO:0042403 |
thyroid hormone metabolic process(GO:0042403) |
| 0.1 |
1.0 |
GO:0050718 |
positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.1 |
0.2 |
GO:0006041 |
glucosamine metabolic process(GO:0006041) |
| 0.1 |
0.2 |
GO:0051231 |
mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) |
| 0.1 |
0.4 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.1 |
1.1 |
GO:0051145 |
smooth muscle cell differentiation(GO:0051145) |
| 0.1 |
0.2 |
GO:1903961 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.1 |
0.4 |
GO:2000124 |
cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 |
0.2 |
GO:0070245 |
positive regulation of T cell apoptotic process(GO:0070234) positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 |
1.3 |
GO:0009214 |
cyclic nucleotide catabolic process(GO:0009214) |
| 0.1 |
0.9 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 |
0.1 |
GO:0036037 |
CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 |
1.3 |
GO:0033144 |
negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) |
| 0.1 |
0.1 |
GO:0019087 |
transformation of host cell by virus(GO:0019087) |
| 0.1 |
0.5 |
GO:0030099 |
myeloid cell differentiation(GO:0030099) |
| 0.1 |
0.4 |
GO:0009051 |
pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 |
1.0 |
GO:0030260 |
entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.1 |
0.4 |
GO:0060999 |
positive regulation of dendritic spine development(GO:0060999) |
| 0.1 |
0.6 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 |
0.1 |
GO:0045885 |
obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.1 |
6.0 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.1 |
0.3 |
GO:0032526 |
response to retinoic acid(GO:0032526) |
| 0.1 |
0.4 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) |
| 0.1 |
0.6 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.1 |
1.1 |
GO:0070206 |
protein trimerization(GO:0070206) |
| 0.1 |
0.2 |
GO:0006307 |
DNA dealkylation involved in DNA repair(GO:0006307) DNA dealkylation(GO:0035510) |
| 0.1 |
1.5 |
GO:0042554 |
superoxide anion generation(GO:0042554) |
| 0.1 |
2.2 |
GO:0060350 |
endochondral bone morphogenesis(GO:0060350) |
| 0.1 |
0.1 |
GO:0042637 |
catagen(GO:0042637) |
| 0.1 |
0.4 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.1 |
0.4 |
GO:0001782 |
B cell homeostasis(GO:0001782) |
| 0.1 |
0.2 |
GO:0060397 |
JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 |
0.2 |
GO:0032581 |
ER-dependent peroxisome organization(GO:0032581) |
| 0.1 |
0.6 |
GO:0034260 |
negative regulation of GTPase activity(GO:0034260) |
| 0.1 |
0.2 |
GO:0017144 |
drug metabolic process(GO:0017144) |
| 0.1 |
0.8 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.1 |
0.1 |
GO:0021859 |
pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.1 |
0.8 |
GO:0048661 |
positive regulation of smooth muscle cell proliferation(GO:0048661) |
| 0.1 |
0.2 |
GO:0034770 |
histone H4-K20 methylation(GO:0034770) |
| 0.1 |
0.8 |
GO:0032456 |
endocytic recycling(GO:0032456) |
| 0.1 |
0.2 |
GO:0018874 |
benzoate metabolic process(GO:0018874) |
| 0.1 |
0.4 |
GO:0051383 |
kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.1 |
0.1 |
GO:0032717 |
negative regulation of interleukin-8 production(GO:0032717) |
| 0.1 |
0.1 |
GO:0051147 |
regulation of muscle cell differentiation(GO:0051147) |
| 0.1 |
0.2 |
GO:0045445 |
myoblast differentiation(GO:0045445) |
| 0.1 |
4.6 |
GO:0008277 |
regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 |
0.5 |
GO:0019614 |
phenol-containing compound catabolic process(GO:0019336) catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.1 |
0.4 |
GO:0010887 |
negative regulation of cholesterol storage(GO:0010887) |
| 0.1 |
0.2 |
GO:0051088 |
PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 |
0.2 |
GO:0051102 |
DNA ligation involved in DNA recombination(GO:0051102) |
| 0.1 |
0.3 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.1 |
2.3 |
GO:0071377 |
cellular response to glucagon stimulus(GO:0071377) |
| 0.1 |
0.3 |
GO:0007281 |
germ cell development(GO:0007281) |
| 0.0 |
0.1 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 |
0.3 |
GO:0006295 |
nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 |
0.1 |
GO:1901522 |
positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 |
0.3 |
GO:2000172 |
regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 |
0.8 |
GO:0030199 |
collagen fibril organization(GO:0030199) |
| 0.0 |
1.2 |
GO:0006911 |
phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 |
0.2 |
GO:0072595 |
maintenance of protein location in nucleus(GO:0051457) maintenance of protein localization in organelle(GO:0072595) |
| 0.0 |
1.0 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.0 |
0.1 |
GO:0016476 |
regulation of embryonic cell shape(GO:0016476) |
| 0.0 |
1.3 |
GO:0018208 |
peptidyl-proline modification(GO:0018208) |
| 0.0 |
0.3 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.0 |
0.5 |
GO:0006098 |
pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 |
0.4 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.0 |
1.0 |
GO:0043101 |
purine-containing compound salvage(GO:0043101) |
| 0.0 |
1.0 |
GO:0002504 |
antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 |
0.0 |
GO:0033590 |
response to cobalamin(GO:0033590) |
| 0.0 |
1.1 |
GO:0007030 |
Golgi organization(GO:0007030) |
| 0.0 |
0.2 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.0 |
0.3 |
GO:0043001 |
Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 |
0.7 |
GO:0006613 |
cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 |
0.2 |
GO:0098927 |
early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 |
0.2 |
GO:0010508 |
positive regulation of autophagy(GO:0010508) |
| 0.0 |
0.3 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 |
0.2 |
GO:0051901 |
positive regulation of mitochondrial depolarization(GO:0051901) positive regulation of membrane depolarization(GO:1904181) |
| 0.0 |
1.0 |
GO:0006555 |
methionine metabolic process(GO:0006555) |
| 0.0 |
0.1 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 |
0.2 |
GO:0043045 |
DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 |
0.1 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 |
0.1 |
GO:0030004 |
cellular monovalent inorganic cation homeostasis(GO:0030004) |
| 0.0 |
0.7 |
GO:0006891 |
intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 |
0.1 |
GO:0000389 |
mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 |
0.1 |
GO:0032926 |
negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 |
0.1 |
GO:0048861 |
leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 |
0.2 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.0 |
0.1 |
GO:0032922 |
circadian regulation of gene expression(GO:0032922) |
| 0.0 |
0.3 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.0 |
0.2 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.0 |
0.1 |
GO:0003351 |
epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 |
0.1 |
GO:0006669 |
sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 |
0.3 |
GO:0051927 |
obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.0 |
0.1 |
GO:0031133 |
regulation of axon diameter(GO:0031133) regulation of cell projection size(GO:0032536) |
| 0.0 |
0.5 |
GO:0048207 |
vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 |
0.2 |
GO:0009439 |
cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.0 |
0.3 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 |
0.9 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 |
0.1 |
GO:0009642 |
response to light intensity(GO:0009642) |
| 0.0 |
0.0 |
GO:0006296 |
nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 |
1.1 |
GO:0080171 |
lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 |
0.2 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 |
0.2 |
GO:0031122 |
cytoplasmic microtubule organization(GO:0031122) |
| 0.0 |
1.0 |
GO:0042531 |
positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 |
0.4 |
GO:0032964 |
collagen biosynthetic process(GO:0032964) |
| 0.0 |
0.2 |
GO:0060261 |
positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 |
0.1 |
GO:0033119 |
negative regulation of RNA splicing(GO:0033119) |
| 0.0 |
1.0 |
GO:0045740 |
positive regulation of DNA replication(GO:0045740) |
| 0.0 |
0.0 |
GO:0050968 |
thermoception(GO:0050955) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 |
0.1 |
GO:0046607 |
positive regulation of centrosome cycle(GO:0046607) |
| 0.0 |
0.3 |
GO:0007413 |
axonal fasciculation(GO:0007413) |
| 0.0 |
0.3 |
GO:0034643 |
establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 |
0.2 |
GO:0032786 |
positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 |
2.2 |
GO:0051781 |
positive regulation of cell division(GO:0051781) |
| 0.0 |
0.1 |
GO:0046341 |
CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 |
0.4 |
GO:0014003 |
oligodendrocyte development(GO:0014003) |
| 0.0 |
0.1 |
GO:0034625 |
fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 |
0.7 |
GO:0006972 |
hyperosmotic response(GO:0006972) |
| 0.0 |
0.0 |
GO:0070167 |
regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 |
0.4 |
GO:0008635 |
activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 |
0.1 |
GO:0006004 |
fucose metabolic process(GO:0006004) |
| 0.0 |
0.1 |
GO:0048712 |
negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 |
0.4 |
GO:0006903 |
vesicle targeting(GO:0006903) |
| 0.0 |
0.1 |
GO:0050774 |
negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 |
0.1 |
GO:0045040 |
outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) establishment of protein localization to mitochondrial membrane(GO:0090151) |
| 0.0 |
0.1 |
GO:0032988 |
ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 |
0.2 |
GO:0045540 |
regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 |
0.7 |
GO:0048678 |
response to axon injury(GO:0048678) |
| 0.0 |
3.0 |
GO:0002576 |
platelet degranulation(GO:0002576) |
| 0.0 |
0.1 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 |
0.1 |
GO:0061084 |
regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) regulation of protein folding(GO:1903332) negative regulation of protein folding(GO:1903333) |
| 0.0 |
0.3 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 |
0.4 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.1 |
GO:0043392 |
negative regulation of DNA binding(GO:0043392) |
| 0.0 |
0.2 |
GO:1900087 |
traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle phase transition(GO:1901989) positive regulation of mitotic cell cycle phase transition(GO:1901992) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.0 |
0.1 |
GO:0003070 |
age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) |
| 0.0 |
0.4 |
GO:0032480 |
negative regulation of type I interferon production(GO:0032480) |
| 0.0 |
0.1 |
GO:0001552 |
ovarian follicle atresia(GO:0001552) |
| 0.0 |
0.1 |
GO:0048295 |
positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 |
0.1 |
GO:0030889 |
negative regulation of B cell proliferation(GO:0030889) |
| 0.0 |
0.1 |
GO:0010815 |
bradykinin catabolic process(GO:0010815) |
| 0.0 |
0.1 |
GO:0031643 |
positive regulation of myelination(GO:0031643) |
| 0.0 |
0.1 |
GO:0051546 |
keratinocyte migration(GO:0051546) |
| 0.0 |
0.1 |
GO:0018101 |
protein citrullination(GO:0018101) citrulline biosynthetic process(GO:0019240) |
| 0.0 |
0.4 |
GO:0045604 |
regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 |
0.1 |
GO:0021836 |
chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.0 |
0.0 |
GO:0042733 |
embryonic digit morphogenesis(GO:0042733) |
| 0.0 |
0.3 |
GO:0035308 |
negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 |
0.2 |
GO:0006998 |
nuclear envelope organization(GO:0006998) |
| 0.0 |
0.0 |
GO:0090292 |
nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 |
0.2 |
GO:0032735 |
positive regulation of interleukin-12 production(GO:0032735) |
| 0.0 |
0.0 |
GO:0043628 |
ncRNA 3'-end processing(GO:0043628) |
| 0.0 |
0.2 |
GO:0048265 |
response to pain(GO:0048265) |
| 0.0 |
0.2 |
GO:0060048 |
cardiac muscle contraction(GO:0060048) |
| 0.0 |
0.1 |
GO:0007035 |
vacuolar acidification(GO:0007035) |
| 0.0 |
0.1 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 |
0.2 |
GO:0070059 |
intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 |
0.3 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.4 |
GO:0006656 |
phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 |
1.2 |
GO:0030833 |
regulation of actin filament polymerization(GO:0030833) |
| 0.0 |
0.2 |
GO:0071157 |
negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 |
0.8 |
GO:0090263 |
positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 |
0.2 |
GO:0007141 |
male meiosis I(GO:0007141) |
| 0.0 |
0.1 |
GO:0048296 |
isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 |
0.1 |
GO:0060124 |
positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 |
0.0 |
GO:0032627 |
interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 |
0.0 |
GO:0060054 |
positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 |
0.2 |
GO:0046831 |
regulation of RNA export from nucleus(GO:0046831) |
| 0.0 |
0.7 |
GO:0007498 |
mesoderm development(GO:0007498) |
| 0.0 |
0.1 |
GO:0042635 |
positive regulation of hair cycle(GO:0042635) positive regulation of epidermis development(GO:0045684) positive regulation of hair follicle development(GO:0051798) |
| 0.0 |
0.2 |
GO:0006734 |
NADH metabolic process(GO:0006734) |
| 0.0 |
0.7 |
GO:1900181 |
negative regulation of protein import into nucleus(GO:0042308) negative regulation of protein localization to nucleus(GO:1900181) negative regulation of protein import(GO:1904590) |
| 0.0 |
0.0 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 |
0.1 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.0 |
0.1 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.0 |
0.2 |
GO:0007519 |
skeletal muscle tissue development(GO:0007519) skeletal muscle organ development(GO:0060538) |
| 0.0 |
0.4 |
GO:0003407 |
neural retina development(GO:0003407) |
| 0.0 |
0.2 |
GO:0006906 |
vesicle fusion(GO:0006906) |
| 0.0 |
0.1 |
GO:0048147 |
negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 |
0.4 |
GO:0061077 |
chaperone-mediated protein folding(GO:0061077) |
| 0.0 |
0.1 |
GO:0042987 |
amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 |
0.2 |
GO:0000209 |
protein polyubiquitination(GO:0000209) |
| 0.0 |
0.1 |
GO:0043114 |
regulation of vascular permeability(GO:0043114) |
| 0.0 |
0.1 |
GO:0006896 |
Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 |
0.3 |
GO:0045648 |
positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 |
0.1 |
GO:0006655 |
phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 |
0.1 |
GO:0042986 |
positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 |
0.1 |
GO:0008089 |
anterograde axonal transport(GO:0008089) axonal transport(GO:0098930) |
| 0.0 |
0.1 |
GO:0006264 |
mitochondrial DNA replication(GO:0006264) |
| 0.0 |
0.4 |
GO:0033540 |
fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 |
0.3 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.0 |
0.0 |
GO:0016198 |
axon choice point recognition(GO:0016198) |
| 0.0 |
3.0 |
GO:0031018 |
endocrine pancreas development(GO:0031018) |
| 0.0 |
0.4 |
GO:0009395 |
phospholipid catabolic process(GO:0009395) |
| 0.0 |
0.3 |
GO:0016197 |
endosomal transport(GO:0016197) |
| 0.0 |
0.1 |
GO:0032677 |
regulation of interleukin-8 production(GO:0032677) |
| 0.0 |
0.5 |
GO:0008154 |
actin polymerization or depolymerization(GO:0008154) |
| 0.0 |
0.0 |
GO:0045065 |
cytotoxic T cell differentiation(GO:0045065) |
| 0.0 |
0.3 |
GO:0030032 |
lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 |
0.1 |
GO:0060155 |
platelet dense granule organization(GO:0060155) |
| 0.0 |
0.1 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.0 |
0.1 |
GO:0046487 |
glyoxylate metabolic process(GO:0046487) |
| 0.0 |
0.0 |
GO:0043653 |
mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 |
0.5 |
GO:0050690 |
regulation of defense response to virus by virus(GO:0050690) |
| 0.0 |
0.4 |
GO:0015949 |
nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 |
0.0 |
GO:0015793 |
glycerol transport(GO:0015793) |
| 0.0 |
0.1 |
GO:0090042 |
tubulin deacetylation(GO:0090042) |
| 0.0 |
0.1 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.0 |
0.1 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 |
0.0 |
GO:0019062 |
virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.0 |
0.1 |
GO:0006477 |
protein sulfation(GO:0006477) |
| 0.0 |
0.1 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.0 |
0.5 |
GO:0019722 |
calcium-mediated signaling(GO:0019722) |
| 0.0 |
0.3 |
GO:0006493 |
protein O-linked glycosylation(GO:0006493) |
| 0.0 |
0.0 |
GO:0045060 |
negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 |
0.2 |
GO:0000768 |
syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 |
0.0 |
GO:0071801 |
podosome assembly(GO:0071800) regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 |
0.2 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.0 |
0.1 |
GO:0044060 |
regulation of endocrine process(GO:0044060) |
| 0.0 |
0.3 |
GO:0008543 |
fibroblast growth factor receptor signaling pathway(GO:0008543) cellular response to fibroblast growth factor stimulus(GO:0044344) response to fibroblast growth factor(GO:0071774) |
| 0.0 |
0.3 |
GO:0043462 |
regulation of ATPase activity(GO:0043462) |
| 0.0 |
0.0 |
GO:0002431 |
Fc receptor mediated stimulatory signaling pathway(GO:0002431) regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 |
0.2 |
GO:0007032 |
endosome organization(GO:0007032) |
| 0.0 |
0.2 |
GO:0031114 |
regulation of microtubule depolymerization(GO:0031114) |
| 0.0 |
0.2 |
GO:0018198 |
peptidyl-cysteine modification(GO:0018198) |
| 0.0 |
0.1 |
GO:0010799 |
regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 |
0.1 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.0 |
0.1 |
GO:0018106 |
peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 |
0.3 |
GO:0007033 |
vacuole organization(GO:0007033) |
| 0.0 |
0.0 |
GO:0000733 |
DNA strand renaturation(GO:0000733) |
| 0.0 |
0.0 |
GO:0070601 |
centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 |
0.0 |
GO:0045872 |
positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 |
0.3 |
GO:0015813 |
L-glutamate transport(GO:0015813) |
| 0.0 |
0.2 |
GO:0035115 |
embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 |
0.0 |
GO:0002667 |
lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 |
0.2 |
GO:0030177 |
positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.0 |
1.0 |
GO:0043087 |
regulation of GTPase activity(GO:0043087) |
| 0.0 |
0.1 |
GO:0051972 |
regulation of telomerase activity(GO:0051972) |
| 0.0 |
0.1 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 |
0.1 |
GO:0000038 |
very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 |
2.0 |
GO:0007389 |
pattern specification process(GO:0007389) |
| 0.0 |
0.1 |
GO:0001539 |
cilium or flagellum-dependent cell motility(GO:0001539) |
| 0.0 |
0.0 |
GO:0007538 |
primary sex determination(GO:0007538) |
| 0.0 |
0.2 |
GO:0001843 |
neural tube closure(GO:0001843) primary neural tube formation(GO:0014020) tube closure(GO:0060606) |
| 0.0 |
0.0 |
GO:0010761 |
fibroblast migration(GO:0010761) |
| 0.0 |
0.6 |
GO:0043473 |
pigmentation(GO:0043473) |
| 0.0 |
0.1 |
GO:0051496 |
positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 |
0.0 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.0 |
0.1 |
GO:0033504 |
floor plate development(GO:0033504) |
| 0.0 |
0.1 |
GO:1901797 |
negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.0 |
0.1 |
GO:0010390 |
histone monoubiquitination(GO:0010390) |
| 0.0 |
0.0 |
GO:0060395 |
SMAD protein signal transduction(GO:0060395) |
| 0.0 |
0.0 |
GO:0001992 |
regulation of systemic arterial blood pressure by vasopressin(GO:0001992) regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.0 |
0.0 |
GO:0070427 |
cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway(GO:0035872) nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 |
1.6 |
GO:0043413 |
protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 |
0.0 |
GO:0060314 |
regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 |
0.4 |
GO:0006029 |
proteoglycan metabolic process(GO:0006029) |
| 0.0 |
0.4 |
GO:0060271 |
cilium morphogenesis(GO:0060271) |
| 0.0 |
0.3 |
GO:0006888 |
ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 |
0.4 |
GO:0007585 |
respiratory gaseous exchange(GO:0007585) |
| 0.0 |
0.3 |
GO:0008344 |
adult locomotory behavior(GO:0008344) |