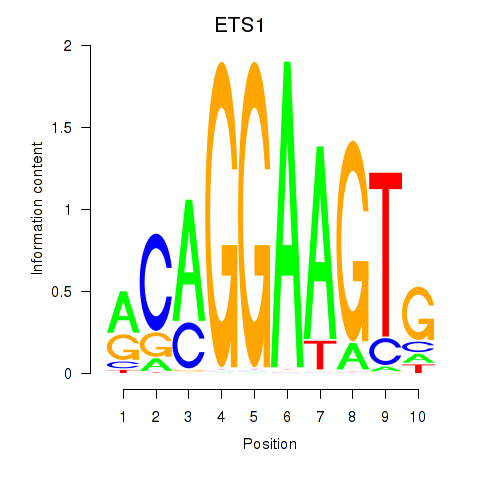
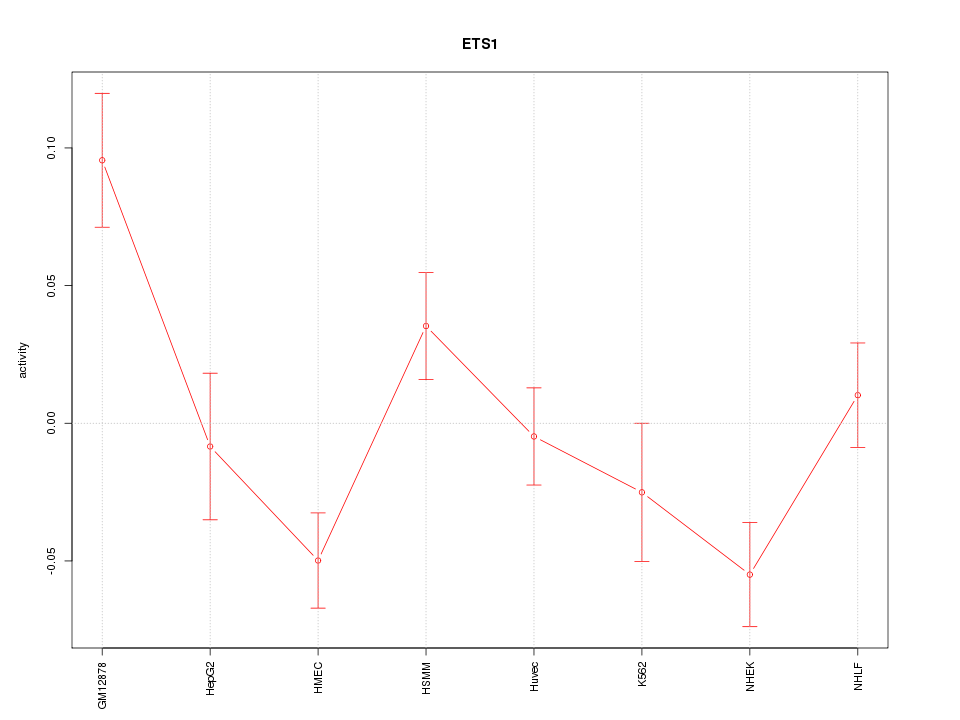
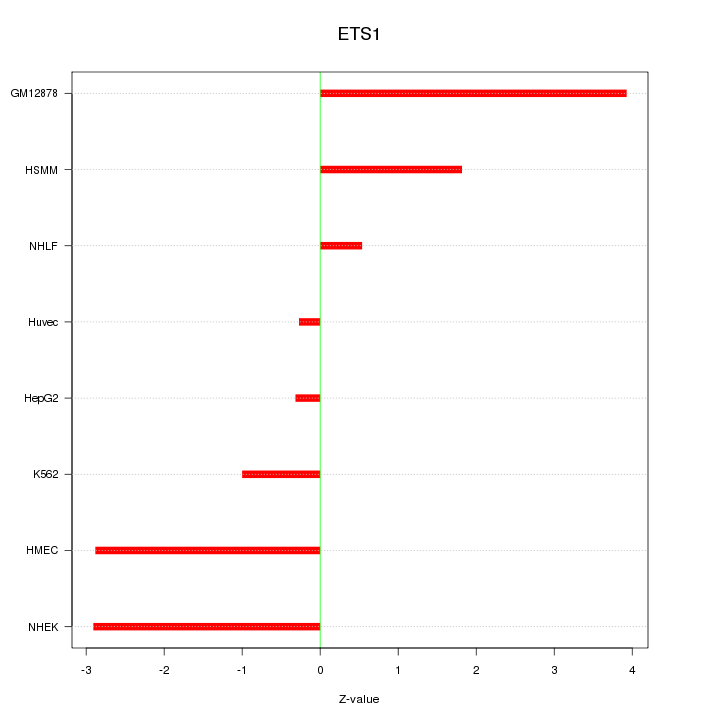
| 1.1 |
4.3 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 1.0 |
2.9 |
GO:0002291 |
T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.9 |
7.3 |
GO:0050857 |
positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.8 |
1.7 |
GO:0009146 |
purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleotide catabolic process(GO:0009155) deoxyribonucleoside triphosphate catabolic process(GO:0009204) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) |
| 0.8 |
3.9 |
GO:0003093 |
regulation of glomerular filtration(GO:0003093) glomerular filtration(GO:0003094) renal filtration(GO:0097205) regulation of renal system process(GO:0098801) |
| 0.6 |
1.9 |
GO:0071380 |
cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.6 |
4.3 |
GO:0034616 |
response to laminar fluid shear stress(GO:0034616) |
| 0.6 |
1.7 |
GO:0019731 |
antibacterial humoral response(GO:0019731) |
| 0.5 |
1.0 |
GO:0070483 |
negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) detection of hypoxia(GO:0070483) |
| 0.5 |
1.4 |
GO:1903961 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.4 |
1.8 |
GO:0033622 |
integrin activation(GO:0033622) regulation of integrin activation(GO:0033623) positive regulation of integrin activation(GO:0033625) |
| 0.4 |
1.3 |
GO:0010836 |
negative regulation of protein ADP-ribosylation(GO:0010836) negative regulation of protein glycosylation(GO:0060051) |
| 0.4 |
1.3 |
GO:0010273 |
regulation of oxidative phosphorylation(GO:0002082) detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.4 |
5.8 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.4 |
1.6 |
GO:0002313 |
mature B cell differentiation involved in immune response(GO:0002313) |
| 0.4 |
2.7 |
GO:0043615 |
astrocyte cell migration(GO:0043615) |
| 0.4 |
1.8 |
GO:0046642 |
negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.4 |
6.3 |
GO:0006906 |
vesicle fusion(GO:0006906) |
| 0.3 |
2.7 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.3 |
1.0 |
GO:0050983 |
spermidine catabolic process(GO:0046203) deoxyhypusine biosynthetic process from spermidine(GO:0050983) |
| 0.3 |
0.8 |
GO:0032328 |
alanine transport(GO:0032328) |
| 0.3 |
1.3 |
GO:0048304 |
positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.2 |
0.7 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.2 |
0.9 |
GO:0010761 |
fibroblast migration(GO:0010761) |
| 0.2 |
1.3 |
GO:0000729 |
DNA double-strand break processing(GO:0000729) |
| 0.2 |
0.4 |
GO:0002335 |
mature B cell differentiation(GO:0002335) |
| 0.2 |
0.6 |
GO:0045601 |
regulation of endothelial cell differentiation(GO:0045601) |
| 0.2 |
0.6 |
GO:1904036 |
negative regulation of epithelial cell apoptotic process(GO:1904036) negative regulation of endothelial cell apoptotic process(GO:2000352) |
| 0.2 |
0.8 |
GO:0002331 |
immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.2 |
2.4 |
GO:0060324 |
face development(GO:0060324) |
| 0.2 |
1.2 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.2 |
1.0 |
GO:0042535 |
positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.2 |
0.6 |
GO:0071460 |
cellular response to cell-matrix adhesion(GO:0071460) |
| 0.2 |
1.8 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.2 |
4.2 |
GO:0050732 |
negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) |
| 0.2 |
0.2 |
GO:0050653 |
chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.2 |
0.5 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) cytoplasmic translation(GO:0002181) cytoplasmic translational initiation(GO:0002183) |
| 0.2 |
0.2 |
GO:0044380 |
protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.2 |
1.0 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.2 |
2.4 |
GO:0050930 |
induction of positive chemotaxis(GO:0050930) |
| 0.2 |
0.8 |
GO:0043923 |
positive regulation by host of viral transcription(GO:0043923) |
| 0.2 |
3.8 |
GO:0050853 |
B cell receptor signaling pathway(GO:0050853) |
| 0.2 |
1.1 |
GO:0060363 |
cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.2 |
1.1 |
GO:0042483 |
negative regulation of odontogenesis(GO:0042483) |
| 0.2 |
0.6 |
GO:0071436 |
sodium ion export(GO:0071436) |
| 0.2 |
1.4 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.2 |
1.4 |
GO:0051973 |
positive regulation of telomerase activity(GO:0051973) |
| 0.2 |
1.2 |
GO:0051383 |
kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.2 |
0.8 |
GO:0043402 |
glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 |
0.6 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.1 |
5.2 |
GO:0030593 |
neutrophil chemotaxis(GO:0030593) neutrophil migration(GO:1990266) |
| 0.1 |
0.9 |
GO:0006658 |
phosphatidylserine metabolic process(GO:0006658) |
| 0.1 |
0.7 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 |
0.7 |
GO:0002726 |
positive regulation of T cell cytokine production(GO:0002726) |
| 0.1 |
2.2 |
GO:0045885 |
obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.1 |
1.1 |
GO:0001782 |
B cell homeostasis(GO:0001782) |
| 0.1 |
1.1 |
GO:0071353 |
cellular response to interleukin-4(GO:0071353) |
| 0.1 |
0.4 |
GO:0009162 |
deoxyribonucleoside monophosphate metabolic process(GO:0009162) pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) transformation of host cell by virus(GO:0019087) dUMP metabolic process(GO:0046078) intestinal epithelial cell maturation(GO:0060574) intestinal epithelial cell development(GO:0060576) |
| 0.1 |
0.4 |
GO:0060744 |
thelarche(GO:0042695) development of secondary female sexual characteristics(GO:0046543) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 |
0.4 |
GO:0060397 |
JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.1 |
0.1 |
GO:0042036 |
negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 |
0.3 |
GO:0021896 |
forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) |
| 0.1 |
0.7 |
GO:0032897 |
negative regulation of viral transcription(GO:0032897) |
| 0.1 |
1.5 |
GO:0071044 |
histone mRNA catabolic process(GO:0071044) |
| 0.1 |
3.8 |
GO:0002286 |
T cell activation involved in immune response(GO:0002286) |
| 0.1 |
0.8 |
GO:0035964 |
COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 |
0.3 |
GO:0031167 |
rRNA methylation(GO:0031167) |
| 0.1 |
1.4 |
GO:0045540 |
regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.1 |
0.4 |
GO:0019276 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 |
0.7 |
GO:0031122 |
cytoplasmic microtubule organization(GO:0031122) |
| 0.1 |
0.5 |
GO:0032007 |
negative regulation of TOR signaling(GO:0032007) |
| 0.1 |
0.5 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.1 |
1.6 |
GO:0032608 |
interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) |
| 0.1 |
0.3 |
GO:0051088 |
PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 |
3.9 |
GO:0050690 |
regulation of defense response to virus by virus(GO:0050690) |
| 0.1 |
0.2 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 |
0.4 |
GO:0010957 |
negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.1 |
0.6 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) |
| 0.1 |
1.8 |
GO:0042531 |
regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 |
0.3 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.1 |
3.8 |
GO:0030183 |
B cell differentiation(GO:0030183) |
| 0.1 |
0.4 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 |
0.4 |
GO:0060528 |
secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 |
1.2 |
GO:0048207 |
vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.1 |
0.3 |
GO:0017014 |
protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.1 |
7.7 |
GO:0050852 |
T cell receptor signaling pathway(GO:0050852) |
| 0.1 |
1.1 |
GO:0051103 |
DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 |
4.0 |
GO:0042113 |
B cell activation(GO:0042113) |
| 0.1 |
0.4 |
GO:0031274 |
pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 |
1.4 |
GO:0006929 |
substrate-dependent cell migration(GO:0006929) |
| 0.1 |
1.2 |
GO:0002920 |
regulation of humoral immune response(GO:0002920) |
| 0.1 |
0.3 |
GO:0090267 |
positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 |
0.3 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 |
0.8 |
GO:0008105 |
asymmetric protein localization(GO:0008105) |
| 0.1 |
0.7 |
GO:0034260 |
negative regulation of GTPase activity(GO:0034260) |
| 0.1 |
0.1 |
GO:2000573 |
positive regulation of DNA biosynthetic process(GO:2000573) |
| 0.1 |
0.5 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
0.2 |
GO:0006110 |
regulation of glycolytic process(GO:0006110) regulation of nucleoside metabolic process(GO:0009118) regulation of ATP metabolic process(GO:1903578) |
| 0.1 |
1.3 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.1 |
0.3 |
GO:0032497 |
detection of lipopolysaccharide(GO:0032497) |
| 0.1 |
0.3 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.1 |
1.9 |
GO:0001756 |
somitogenesis(GO:0001756) |
| 0.1 |
0.4 |
GO:0071459 |
protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 |
0.2 |
GO:0018283 |
metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.1 |
0.7 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 |
0.6 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.1 |
0.3 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 |
0.3 |
GO:0007035 |
vacuolar acidification(GO:0007035) |
| 0.1 |
0.5 |
GO:0032703 |
negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 |
1.2 |
GO:0006662 |
glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 |
0.2 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 |
0.4 |
GO:0045003 |
DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 |
0.0 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.0 |
0.4 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.0 |
1.5 |
GO:0030866 |
cortical cytoskeleton organization(GO:0030865) cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 |
0.5 |
GO:0047496 |
vesicle transport along microtubule(GO:0047496) vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 |
0.1 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) response to cobalt ion(GO:0032025) positive regulation of sarcomere organization(GO:0060298) |
| 0.0 |
0.2 |
GO:0035090 |
maintenance of cell polarity(GO:0030011) maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 |
0.3 |
GO:0006553 |
lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 |
0.4 |
GO:0006975 |
DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 |
1.1 |
GO:0051225 |
spindle assembly(GO:0051225) |
| 0.0 |
0.2 |
GO:0032516 |
positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 |
0.1 |
GO:0003351 |
epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 |
0.1 |
GO:0045065 |
cytotoxic T cell differentiation(GO:0045065) |
| 0.0 |
0.3 |
GO:0042474 |
middle ear morphogenesis(GO:0042474) |
| 0.0 |
0.4 |
GO:0031293 |
membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 |
0.3 |
GO:0002090 |
regulation of receptor internalization(GO:0002090) positive regulation of receptor internalization(GO:0002092) |
| 0.0 |
0.6 |
GO:0051319 |
mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 |
0.2 |
GO:1904893 |
negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 |
0.3 |
GO:0034405 |
response to fluid shear stress(GO:0034405) |
| 0.0 |
0.2 |
GO:0042693 |
muscle cell fate commitment(GO:0042693) |
| 0.0 |
1.1 |
GO:0006024 |
glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 |
0.3 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 |
0.3 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 |
0.6 |
GO:0007032 |
endosome organization(GO:0007032) |
| 0.0 |
0.1 |
GO:0032509 |
endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 |
0.2 |
GO:0051123 |
RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 |
0.2 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.0 |
0.2 |
GO:1901985 |
positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 |
0.4 |
GO:0016601 |
Rac protein signal transduction(GO:0016601) |
| 0.0 |
0.3 |
GO:0010815 |
bradykinin catabolic process(GO:0010815) |
| 0.0 |
0.5 |
GO:0006613 |
cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 |
0.0 |
GO:0060684 |
epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 |
0.3 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) histone H4-K12 acetylation(GO:0043983) |
| 0.0 |
0.8 |
GO:0006829 |
zinc II ion transport(GO:0006829) |
| 0.0 |
0.1 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.0 |
0.0 |
GO:0042637 |
catagen(GO:0042637) |
| 0.0 |
0.1 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 |
0.6 |
GO:0008380 |
RNA splicing(GO:0008380) |
| 0.0 |
0.1 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
0.4 |
GO:0000090 |
mitotic anaphase(GO:0000090) |
| 0.0 |
0.3 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
0.2 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.0 |
0.4 |
GO:0045954 |
positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 |
1.6 |
GO:0050680 |
negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 |
0.0 |
GO:0046619 |
optic placode formation(GO:0001743) optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 |
0.2 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
0.1 |
GO:0006651 |
diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 |
0.1 |
GO:0060456 |
positive regulation of digestive system process(GO:0060456) |
| 0.0 |
0.2 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.0 |
0.3 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.0 |
0.3 |
GO:0032876 |
regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 |
0.3 |
GO:0032964 |
collagen biosynthetic process(GO:0032964) |
| 0.0 |
0.5 |
GO:0010648 |
negative regulation of cell communication(GO:0010648) |
| 0.0 |
0.6 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.0 |
0.2 |
GO:0006379 |
mRNA cleavage(GO:0006379) |
| 0.0 |
0.1 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.0 |
2.7 |
GO:0043010 |
camera-type eye development(GO:0043010) |
| 0.0 |
0.3 |
GO:0042787 |
protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 |
0.6 |
GO:0006099 |
tricarboxylic acid cycle(GO:0006099) |
| 0.0 |
0.1 |
GO:0010216 |
maintenance of DNA methylation(GO:0010216) |
| 0.0 |
0.6 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
0.6 |
GO:0036503 |
ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) ERAD pathway(GO:0036503) |
| 0.0 |
1.6 |
GO:0007229 |
integrin-mediated signaling pathway(GO:0007229) |
| 0.0 |
0.1 |
GO:0045943 |
positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 |
0.2 |
GO:0046831 |
regulation of RNA export from nucleus(GO:0046831) |
| 0.0 |
0.0 |
GO:0045136 |
development of secondary sexual characteristics(GO:0045136) |
| 0.0 |
0.1 |
GO:0010544 |
negative regulation of platelet activation(GO:0010544) |
| 0.0 |
0.2 |
GO:0071157 |
negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 |
0.6 |
GO:0042771 |
intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 |
1.5 |
GO:0009408 |
response to heat(GO:0009408) |
| 0.0 |
0.7 |
GO:0005980 |
glycogen catabolic process(GO:0005980) |
| 0.0 |
0.1 |
GO:0000114 |
obsolete regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0000114) |
| 0.0 |
1.3 |
GO:0043473 |
pigmentation(GO:0043473) |
| 0.0 |
0.1 |
GO:0043392 |
negative regulation of DNA binding(GO:0043392) |
| 0.0 |
0.1 |
GO:0035019 |
somatic stem cell population maintenance(GO:0035019) |
| 0.0 |
0.2 |
GO:0006903 |
vesicle targeting(GO:0006903) |
| 0.0 |
0.1 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 |
0.0 |
GO:0060155 |
platelet dense granule organization(GO:0060155) |
| 0.0 |
0.0 |
GO:0045618 |
positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 |
0.3 |
GO:0042994 |
cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 |
0.0 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 |
0.1 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.2 |
GO:0006884 |
cell volume homeostasis(GO:0006884) |
| 0.0 |
0.5 |
GO:0051318 |
G1 phase(GO:0051318) |
| 0.0 |
0.9 |
GO:0051092 |
positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 |
4.2 |
GO:0016337 |
single organismal cell-cell adhesion(GO:0016337) |
| 0.0 |
0.1 |
GO:0001885 |
endothelial cell development(GO:0001885) |
| 0.0 |
0.2 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
0.2 |
GO:0010390 |
histone monoubiquitination(GO:0010390) |
| 0.0 |
0.1 |
GO:0032506 |
barrier septum assembly(GO:0000917) cytokinetic process(GO:0032506) cell septum assembly(GO:0090529) mitotic cytokinetic process(GO:1902410) |
| 0.0 |
1.7 |
GO:0065004 |
protein-DNA complex assembly(GO:0065004) |
| 0.0 |
0.3 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.2 |
GO:0045648 |
positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 |
1.2 |
GO:0007059 |
chromosome segregation(GO:0007059) |
| 0.0 |
0.6 |
GO:0030071 |
regulation of mitotic metaphase/anaphase transition(GO:0030071) regulation of metaphase/anaphase transition of cell cycle(GO:1902099) |
| 0.0 |
0.1 |
GO:0010821 |
regulation of mitochondrion organization(GO:0010821) |
| 0.0 |
0.1 |
GO:0045040 |
outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 |
0.1 |
GO:0060123 |
regulation of growth hormone secretion(GO:0060123) positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 |
0.1 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 |
0.3 |
GO:0006297 |
nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 |
0.3 |
GO:0008214 |
protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 |
0.2 |
GO:0007131 |
reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 |
0.6 |
GO:0001764 |
neuron migration(GO:0001764) |
| 0.0 |
0.4 |
GO:0018208 |
peptidyl-proline modification(GO:0018208) |
| 0.0 |
0.0 |
GO:0016082 |
synaptic vesicle priming(GO:0016082) |
| 0.0 |
0.1 |
GO:0090224 |
regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
| 0.0 |
0.2 |
GO:0070830 |
apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 |
0.2 |
GO:0007498 |
mesoderm development(GO:0007498) |
| 0.0 |
1.1 |
GO:0007588 |
excretion(GO:0007588) |
| 0.0 |
0.2 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.8 |
GO:0010827 |
regulation of glucose transport(GO:0010827) |
| 0.0 |
0.2 |
GO:0006825 |
copper ion transport(GO:0006825) |
| 0.0 |
0.1 |
GO:0001936 |
regulation of endothelial cell proliferation(GO:0001936) |
| 0.0 |
0.7 |
GO:0006986 |
response to unfolded protein(GO:0006986) |
| 0.0 |
1.0 |
GO:0008360 |
regulation of cell shape(GO:0008360) |
| 0.0 |
0.5 |
GO:0001837 |
epithelial to mesenchymal transition(GO:0001837) |
| 0.0 |
0.0 |
GO:0006290 |
pyrimidine dimer repair(GO:0006290) |
| 0.0 |
0.2 |
GO:0007281 |
germ cell development(GO:0007281) |
| 0.0 |
0.2 |
GO:0060334 |
regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 |
0.0 |
GO:0043628 |
ncRNA 3'-end processing(GO:0043628) |
| 0.0 |
0.1 |
GO:0045010 |
actin nucleation(GO:0045010) |
| 0.0 |
0.2 |
GO:0008625 |
extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 |
0.2 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.0 |
0.0 |
GO:0051294 |
establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 |
0.1 |
GO:0006362 |
transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 |
0.5 |
GO:0034332 |
adherens junction organization(GO:0034332) |
| 0.0 |
0.1 |
GO:0030838 |
positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 |
0.2 |
GO:0006914 |
autophagy(GO:0006914) |
| 0.0 |
0.2 |
GO:0042776 |
mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 |
0.1 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.0 |
0.2 |
GO:0048009 |
insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 |
0.0 |
GO:0032392 |
DNA geometric change(GO:0032392) |
| 0.0 |
0.2 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
0.0 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.0 |
0.1 |
GO:0040018 |
positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 |
0.3 |
GO:0046320 |
regulation of fatty acid oxidation(GO:0046320) |
| 0.0 |
0.1 |
GO:0007598 |
blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 |
0.2 |
GO:0006414 |
translational elongation(GO:0006414) |