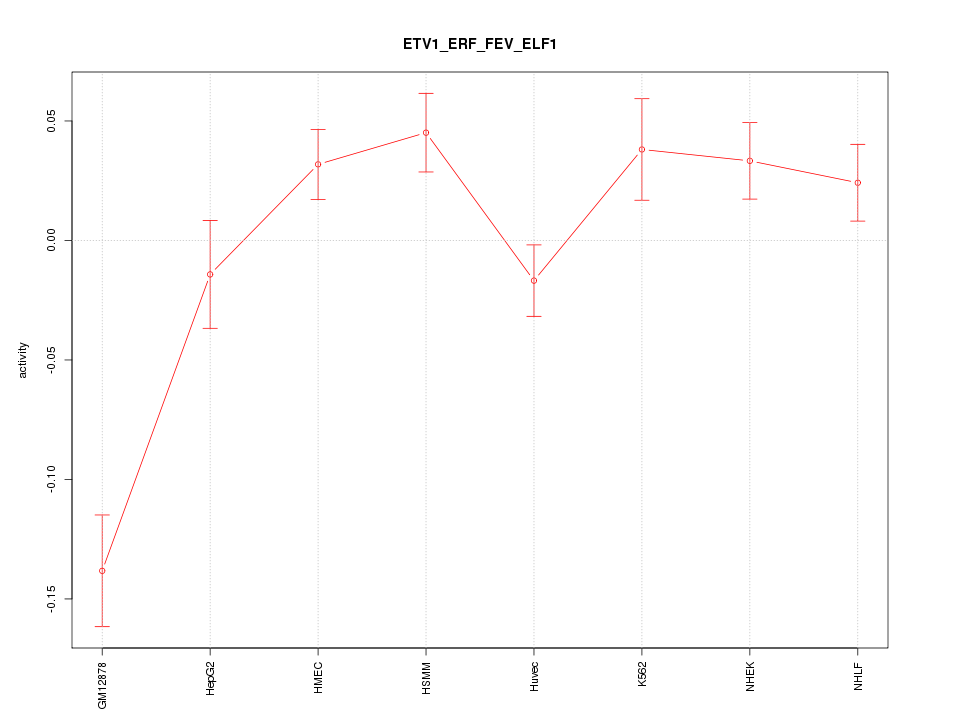
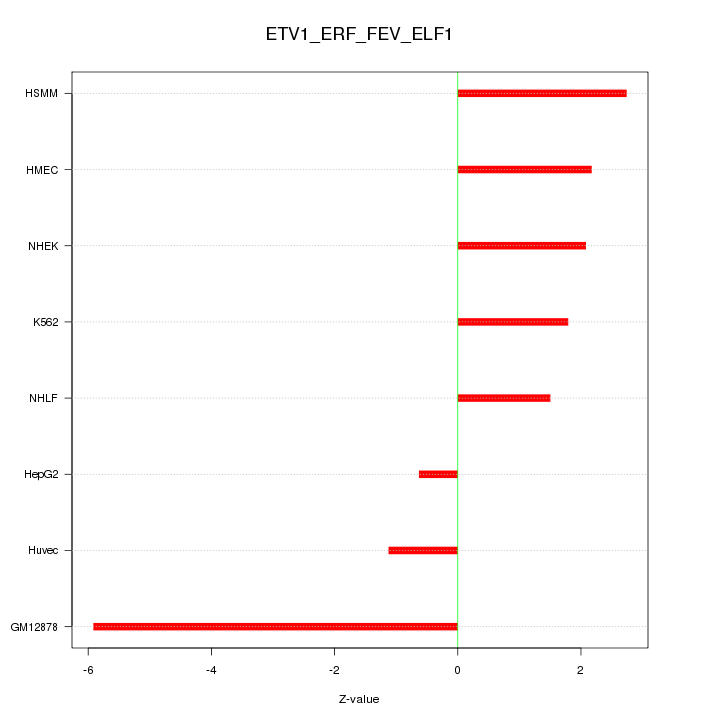
Motif ID: ETV1_ERF_FEV_ELF1
Z-value: 2.709
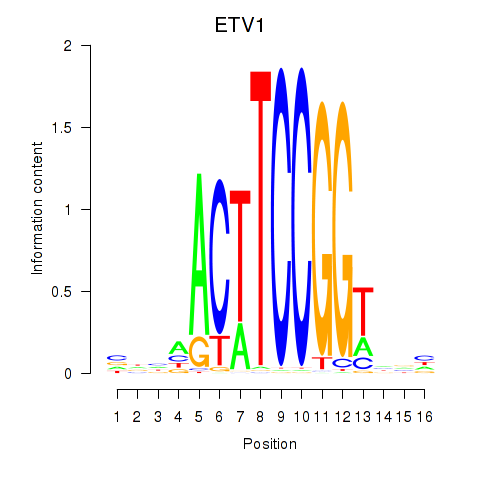
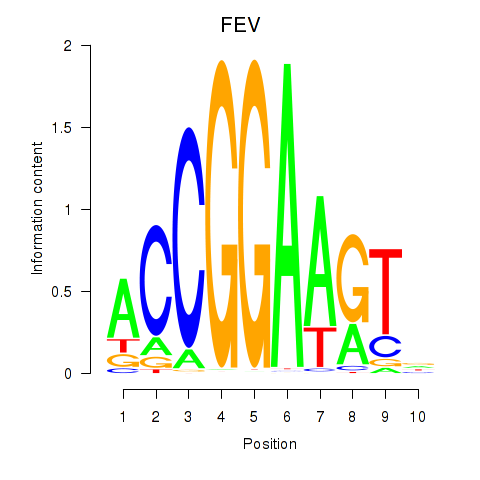
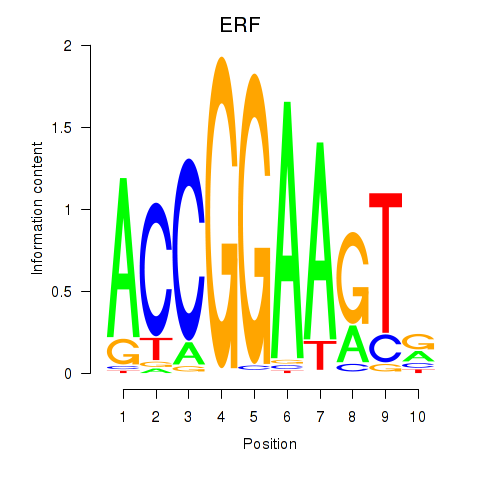
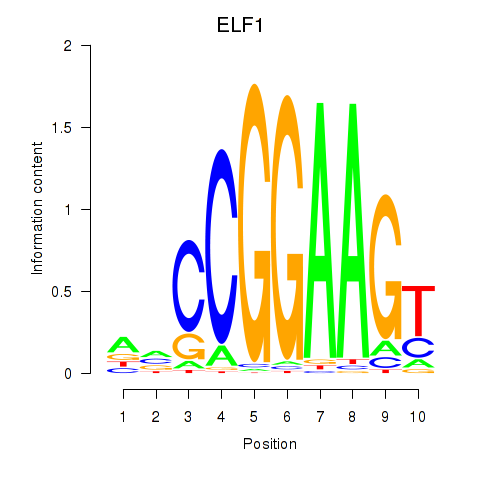
Transcription factors associated with ETV1_ERF_FEV_ELF1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| ELF1 | ENSG00000120690.9 | ELF1 |
| ERF | ENSG00000105722.5 | ERF |
| ETV1 | ENSG00000006468.9 | ETV1 |
| FEV | ENSG00000163497.2 | FEV |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.6 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 2.3 | 18.5 | GO:0003065 | regulation of heart rate by chemical signal(GO:0003062) positive regulation of heart rate by epinephrine(GO:0003065) |
| 1.6 | 4.7 | GO:0035269 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 1.4 | 4.2 | GO:0060295 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) cilium movement involved in cell motility(GO:0060294) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 1.3 | 4.0 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 1.0 | 5.7 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.9 | 2.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.9 | 2.6 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.8 | 3.3 | GO:0071025 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.8 | 8.2 | GO:0046051 | GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.8 | 3.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.7 | 2.0 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.6 | 2.5 | GO:0060209 | estrus(GO:0060209) |
| 0.6 | 1.9 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) Sertoli cell proliferation(GO:0060011) |
| 0.6 | 4.8 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.6 | 0.6 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.6 | 2.3 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.5 | 1.6 | GO:0046368 | GDP-L-fucose biosynthetic process(GO:0042350) 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) GDP-L-fucose metabolic process(GO:0046368) |
| 0.5 | 2.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.5 | 4.7 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.5 | 4.0 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.5 | 1.4 | GO:0016241 | regulation of macroautophagy(GO:0016241) |
| 0.5 | 2.8 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.5 | 3.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.4 | 1.3 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.4 | 1.3 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.4 | 1.8 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.4 | 2.6 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.4 | 2.1 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.4 | 7.5 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.4 | 0.8 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.4 | 5.2 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.4 | 0.4 | GO:0014889 | muscle atrophy(GO:0014889) |
| 0.4 | 3.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.4 | 7.8 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.4 | 1.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.3 | 4.1 | GO:0042987 | amyloid precursor protein catabolic process(GO:0042987) |
| 0.3 | 1.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.3 | 8.4 | GO:0044349 | nucleotide-excision repair, DNA damage removal(GO:0000718) DNA excision(GO:0044349) |
| 0.3 | 3.6 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.3 | 9.1 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.3 | 0.3 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.3 | 2.7 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.3 | 2.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.3 | 4.4 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.3 | 1.9 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.3 | 1.6 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.3 | 0.8 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.3 | 5.6 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.3 | 3.7 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.3 | 3.6 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.3 | 2.0 | GO:0097384 | cellular lipid biosynthetic process(GO:0097384) |
| 0.3 | 21.6 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.2 | 1.7 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.2 | 1.5 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.2 | 3.3 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.2 | 0.7 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.2 | 1.6 | GO:0000239 | pachytene(GO:0000239) |
| 0.2 | 1.2 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.2 | 0.7 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.2 | 0.2 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.2 | 0.9 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 | 1.3 | GO:0032472 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) Golgi calcium ion transport(GO:0032472) manganese ion homeostasis(GO:0055071) |
| 0.2 | 1.5 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.2 | 0.6 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.2 | 10.6 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.2 | 3.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.2 | 8.1 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.2 | 1.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.2 | 1.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.2 | 1.8 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.2 | 3.4 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.2 | 0.6 | GO:0046520 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.2 | 1.2 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.2 | 23.8 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.2 | 0.8 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.2 | 1.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 | 0.9 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 1.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.2 | 3.7 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.2 | 0.5 | GO:0072387 | flavin-containing compound biosynthetic process(GO:0042727) flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.2 | 0.4 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.2 | 0.4 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.2 | 0.9 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.2 | 0.9 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.2 | 6.9 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.2 | 1.1 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.2 | 0.9 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.2 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 0.9 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.2 | 0.5 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.2 | 1.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 0.5 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.2 | 0.7 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.2 | 0.8 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.2 | 0.7 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.2 | 0.5 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) |
| 0.2 | 0.5 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.2 | 2.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 0.8 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.2 | 0.5 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.2 | 0.6 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.2 | 0.5 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) negative regulation of endothelial cell apoptotic process(GO:2000352) |
| 0.2 | 1.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.2 | 0.6 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.2 | 2.1 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.6 | GO:0018126 | protein hydroxylation(GO:0018126) recognition of apoptotic cell(GO:0043654) |
| 0.1 | 1.5 | GO:0006379 | mRNA cleavage(GO:0006379) RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.1 | 0.6 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.1 | 0.4 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 0.7 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.1 | 0.4 | GO:0000917 | barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) mitotic cytokinetic process(GO:1902410) |
| 0.1 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 1.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 1.7 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 1.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.8 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.1 | 1.3 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 1.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.5 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 2.2 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.1 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 2.7 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.1 | 2.4 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 0.5 | GO:0051043 | regulation of membrane protein ectodomain proteolysis(GO:0051043) |
| 0.1 | 0.4 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.2 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.1 | 0.4 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.1 | 0.5 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 0.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 0.4 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 8.2 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.1 | 0.5 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) |
| 0.1 | 0.5 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 3.6 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.1 | 3.1 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.1 | 0.7 | GO:0015840 | urea transport(GO:0015840) |
| 0.1 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 1.6 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.3 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 1.0 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.1 | 0.9 | GO:0043558 | regulation of translational initiation in response to stress(GO:0043558) |
| 0.1 | 2.8 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.4 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 | 2.1 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 0.8 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 7.0 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.1 | 0.8 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.1 | 0.5 | GO:0032042 | mitochondrial DNA replication(GO:0006264) mitochondrial DNA metabolic process(GO:0032042) |
| 0.1 | 0.4 | GO:0000959 | mitochondrial RNA metabolic process(GO:0000959) transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.3 | GO:0046015 | carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.1 | 2.3 | GO:0006490 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 0.5 | GO:0051168 | nuclear export(GO:0051168) |
| 0.1 | 1.0 | GO:1901984 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.1 | 0.2 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.1 | 0.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.4 | GO:0019614 | phenol-containing compound catabolic process(GO:0019336) catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.1 | 0.4 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.6 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.1 | 0.3 | GO:1901799 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) negative regulation of proteasomal protein catabolic process(GO:1901799) negative regulation of proteolysis involved in cellular protein catabolic process(GO:1903051) negative regulation of cellular protein catabolic process(GO:1903363) |
| 0.1 | 0.4 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.1 | 0.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.2 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.1 | 0.3 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.1 | 2.7 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.1 | 0.3 | GO:0060178 | exocyst assembly(GO:0001927) regulation of exocyst assembly(GO:0001928) exocyst localization(GO:0051601) regulation of exocyst localization(GO:0060178) |
| 0.1 | 0.5 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.1 | 0.2 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 3.4 | GO:0033572 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 | 6.9 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 2.8 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.6 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 0.5 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.1 | 3.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 1.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 0.2 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 0.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.1 | 2.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.5 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.1 | 1.1 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 1.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 1.2 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.1 | 0.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 5.9 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.1 | 0.9 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.4 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.9 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 1.1 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.1 | 1.5 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.8 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.4 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.1 | 0.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 1.3 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 1.2 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.1 | 0.8 | GO:0030163 | protein catabolic process(GO:0030163) |
| 0.1 | 0.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 1.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.3 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.3 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.1 | 0.5 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.1 | 1.0 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.1 | 0.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.1 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 1.2 | GO:0000768 | syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.1 | 0.7 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.9 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 0.6 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.1 | 0.6 | GO:1901224 | activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.1 | 0.6 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 2.7 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.1 | GO:0010919 | regulation of inositol phosphate biosynthetic process(GO:0010919) positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.8 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.1 | 1.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.7 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 | 0.3 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.1 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.6 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 1.2 | GO:0008633 | obsolete activation of pro-apoptotic gene products(GO:0008633) |
| 0.1 | 0.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 2.9 | GO:0042594 | response to starvation(GO:0042594) |
| 0.1 | 0.5 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 0.4 | GO:2001021 | negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.1 | 0.6 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.1 | 0.5 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 0.3 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.1 | 0.1 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.1 | 0.3 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.1 | 0.2 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 2.3 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.1 | 6.3 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.0 | 0.0 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.6 | GO:0016458 | gene silencing(GO:0016458) |
| 0.0 | 0.5 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.3 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) regulation of cholesterol homeostasis(GO:2000188) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 1.2 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 2.7 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.0 | GO:0061101 | neuroendocrine cell differentiation(GO:0061101) |
| 0.0 | 0.4 | GO:0001960 | negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.0 | 0.2 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.0 | 0.2 | GO:0090343 | positive regulation of cell aging(GO:0090343) |
| 0.0 | 0.4 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 2.5 | GO:0030336 | negative regulation of cell migration(GO:0030336) negative regulation of cell motility(GO:2000146) |
| 0.0 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 4.2 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.3 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 2.3 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.2 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine development(GO:0060998) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 5.7 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.2 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.5 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 1.8 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.9 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.1 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 1.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 1.2 | GO:0070664 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) negative regulation of leukocyte proliferation(GO:0070664) |
| 0.0 | 1.4 | GO:0001889 | liver development(GO:0001889) |
| 0.0 | 0.7 | GO:0050805 | negative regulation of synaptic transmission(GO:0050805) |
| 0.0 | 2.0 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.4 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 6.3 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.1 | GO:0046604 | regulation of mitotic centrosome separation(GO:0046602) positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.4 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.0 | 1.9 | GO:0008213 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 0.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.5 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 1.0 | GO:0042384 | cilium assembly(GO:0042384) |
| 0.0 | 0.9 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.2 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.0 | 0.1 | GO:0046639 | negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) T-helper 2 cell differentiation(GO:0045064) negative regulation of T-helper cell differentiation(GO:0045623) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of alpha-beta T cell activation(GO:0046636) negative regulation of alpha-beta T cell differentiation(GO:0046639) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.0 | 0.3 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.0 | 1.3 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 1.0 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.2 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.2 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.5 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 2.2 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.3 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 0.4 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.3 | GO:0033158 | regulation of protein import into nucleus, translocation(GO:0033158) |
| 0.0 | 0.2 | GO:0021955 | central nervous system projection neuron axonogenesis(GO:0021952) central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 2.8 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.4 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.3 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.0 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 1.2 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) activation of cysteine-type endopeptidase activity(GO:0097202) |
| 0.0 | 0.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:2000398 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 | 0.5 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.4 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 0.8 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.3 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 0.2 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 0.7 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.2 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.7 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:1900087 | traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle phase transition(GO:1901989) positive regulation of mitotic cell cycle phase transition(GO:1901992) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.0 | 0.5 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 | 0.4 | GO:0010332 | response to gamma radiation(GO:0010332) |
| 0.0 | 1.5 | GO:0007034 | vacuolar transport(GO:0007034) |
| 0.0 | 0.6 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.2 | GO:0038179 | neurotrophin signaling pathway(GO:0038179) neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.9 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.2 | GO:0007632 | visual behavior(GO:0007632) |
| 0.0 | 0.1 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.4 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.4 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.0 | GO:1902803 | regulation of synaptic vesicle transport(GO:1902803) regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.0 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.2 | GO:0000216 | obsolete M/G1 transition of mitotic cell cycle(GO:0000216) |
| 0.0 | 0.9 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.3 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 0.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.9 | GO:0048588 | developmental cell growth(GO:0048588) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.8 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) cellular response to fibroblast growth factor stimulus(GO:0044344) response to fibroblast growth factor(GO:0071774) |
| 0.0 | 0.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.4 | GO:0001824 | blastocyst development(GO:0001824) |
| 0.0 | 1.4 | GO:0034504 | protein localization to nucleus(GO:0034504) |
| 0.0 | 0.8 | GO:0006213 | pyrimidine nucleoside metabolic process(GO:0006213) |
| 0.0 | 0.1 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 | 5.7 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 3.7 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 2.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0009791 | post-embryonic development(GO:0009791) |
| 0.0 | 0.1 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 1.4 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 0.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.9 | GO:0070302 | regulation of stress-activated MAPK cascade(GO:0032872) regulation of stress-activated protein kinase signaling cascade(GO:0070302) |
| 0.0 | 0.2 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.0 | 0.0 | GO:0000189 | MAPK import into nucleus(GO:0000189) pyrimidine nucleobase metabolic process(GO:0006206) cytosine metabolic process(GO:0019858) |
| 0.0 | 0.1 | GO:0072595 | maintenance of protein localization in organelle(GO:0072595) |
| 0.0 | 0.2 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.0 | 0.1 | GO:0006111 | regulation of gluconeogenesis(GO:0006111) |
| 0.0 | 0.2 | GO:0042977 | activation of JAK2 kinase activity(GO:0042977) |
| 0.0 | 0.9 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 2.2 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 0.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.0 | GO:0009299 | mRNA transcription(GO:0009299) mRNA transcription from RNA polymerase II promoter(GO:0042789) determination of dorsal/ventral asymmetry(GO:0048262) determination of dorsal identity(GO:0048263) |
| 0.0 | 0.2 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.3 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.1 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.8 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0001774 | microglial cell activation(GO:0001774) microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.0 | GO:0070781 | response to biotin(GO:0070781) |
| 0.0 | 0.1 | GO:0030260 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.3 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.0 | GO:0015812 | gamma-aminobutyric acid secretion(GO:0014051) regulation of gamma-aminobutyric acid secretion(GO:0014052) gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.0 | GO:0022616 | DNA strand elongation(GO:0022616) |
| 0.0 | 0.2 | GO:1902582 | single-organism intracellular transport(GO:1902582) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 1.0 | GO:0051320 | mitotic S phase(GO:0000084) S phase(GO:0051320) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.1 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.4 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.3 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0042269 | regulation of natural killer cell mediated immunity(GO:0002715) regulation of natural killer cell mediated cytotoxicity(GO:0042269) |
| 0.0 | 0.1 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 1.2 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 3.0 | GO:0007283 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 0.1 | GO:0048265 | response to pain(GO:0048265) |
| 0.0 | 0.0 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.0 | 0.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.2 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 2.0 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 0.1 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.0 | 0.2 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.2 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 0.1 | GO:0051051 | negative regulation of transport(GO:0051051) |
| 0.0 | 1.0 | GO:0006874 | cellular calcium ion homeostasis(GO:0006874) |
| 0.0 | 0.2 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.6 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0031501 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 1.6 | 18.8 | GO:0032059 | bleb(GO:0032059) |
| 1.3 | 7.6 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 1.0 | 5.0 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.9 | 2.7 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.8 | 3.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.8 | 6.9 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.7 | 2.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.7 | 2.7 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.6 | 2.5 | GO:0043219 | lateral loop(GO:0043219) |
| 0.6 | 4.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.6 | 8.0 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.5 | 2.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.5 | 6.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.5 | 4.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.5 | 1.9 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.5 | 2.8 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.5 | 6.3 | GO:0022624 | proteasome accessory complex(GO:0022624) |
| 0.4 | 4.8 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.4 | 5.1 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.4 | 3.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.4 | 1.9 | GO:0032806 | carboxy-terminal domain protein kinase complex(GO:0032806) |
| 0.4 | 4.1 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.4 | 3.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.4 | 1.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.4 | 1.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.4 | 3.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.3 | 8.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.3 | 1.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.3 | 4.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.3 | 1.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.3 | 2.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.3 | 0.9 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.3 | 3.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.3 | 2.0 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.3 | 0.9 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.3 | 4.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.3 | 3.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 2.8 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.2 | 7.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 2.0 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.2 | 1.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.2 | 6.9 | GO:0090568 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.2 | 0.7 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 0.9 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.2 | 0.9 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.2 | 6.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 0.2 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) UBC13-MMS2 complex(GO:0031372) |
| 0.2 | 2.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 2.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.2 | 0.9 | GO:0038201 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.2 | 1.5 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.2 | 4.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 1.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 0.6 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 1.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.2 | 2.0 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 12.5 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.2 | 6.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.2 | 0.9 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.2 | 1.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 2.7 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.2 | 1.9 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.2 | 1.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 0.8 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.2 | 0.5 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 4.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 18.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 0.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 1.9 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 2.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 1.7 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.5 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.5 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 3.2 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.1 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 12.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.8 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.1 | 1.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 5.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 2.0 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 1.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 0.4 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.4 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.1 | 0.7 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.6 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.7 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 6.1 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 2.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 0.5 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 1.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 2.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 1.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 18.2 | GO:0044440 | endosomal part(GO:0044440) |
| 0.1 | 1.0 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 4.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.0 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.5 | GO:0097610 | cell division site(GO:0032153) cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
| 0.1 | 1.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.4 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.1 | 1.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 0.7 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.3 | GO:0032449 | CBM complex(GO:0032449) |
| 0.1 | 0.2 | GO:0014802 | terminal cisterna(GO:0014802) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 6.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 2.0 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.1 | 1.6 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 1.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.5 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.5 | GO:0044452 | nucleolar part(GO:0044452) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 1.3 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.3 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 46.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.1 | GO:1990316 | pre-autophagosomal structure(GO:0000407) ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 1.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.0 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.5 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 2.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 2.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.6 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.6 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 1.1 | GO:0042645 | mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.4 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 1.6 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.0 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 1.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.0 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 1.2 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.8 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 8.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.9 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 3.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.6 | GO:0001725 | stress fiber(GO:0001725) actin filament bundle(GO:0032432) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.5 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 3.2 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 6.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 5.7 | GO:0005625 | obsolete soluble fraction(GO:0005625) |
| 0.0 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.0 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 29.1 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 0.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.7 | GO:0019717 | obsolete synaptosome(GO:0019717) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0019867 | outer membrane(GO:0019867) |
| 0.0 | 0.1 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) |
| 0.0 | 2.3 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0016528 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 1.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.7 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.2 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.1 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.2 | GO:0044431 | Golgi apparatus part(GO:0044431) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.6 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 1.6 | 4.7 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 1.3 | 4.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 1.3 | 3.9 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 1.2 | 3.7 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.9 | 2.7 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.9 | 6.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.8 | 2.5 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.8 | 8.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.7 | 2.9 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.7 | 2.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.6 | 3.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.6 | 3.8 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.6 | 1.9 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.6 | 2.9 | GO:0080030 | methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.5 | 2.0 | GO:0008148 | obsolete negative transcription elongation factor activity(GO:0008148) |
| 0.5 | 2.0 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.5 | 16.2 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.5 | 2.9 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.5 | 12.8 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.4 | 1.3 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.4 | 1.8 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.4 | 3.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.4 | 1.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.4 | 2.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.4 | 5.2 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.4 | 3.7 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.4 | 2.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.4 | 1.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.4 | 1.8 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.4 | 1.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.4 | 3.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.4 | 7.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.3 | 4.4 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.3 | 1.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.3 | 0.6 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.3 | 8.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.3 | 1.5 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.3 | 0.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.3 | 1.5 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.3 | 0.6 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.3 | 2.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.3 | 2.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.3 | 3.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.3 | 1.8 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.3 | 1.3 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.3 | 1.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 0.9 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.2 | 5.9 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.2 | 1.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.2 | 0.7 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.2 | 0.9 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 0.9 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 0.7 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.2 | 1.3 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.2 | 0.9 | GO:0019961 | interferon binding(GO:0019961) |
| 0.2 | 0.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 0.8 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.2 | 4.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.2 | 6.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 19.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 1.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 1.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.2 | 0.8 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 0.8 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.2 | 9.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 0.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) |
| 0.2 | 2.7 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.2 | 0.8 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.2 | 6.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 3.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 1.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.2 | 0.5 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.2 | 1.9 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 0.9 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 1.2 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.2 | 0.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 1.7 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 1.4 | GO:0046934 | obsolete inositol or phosphatidylinositol kinase activity(GO:0004428) phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.2 | 1.7 | GO:0016208 | AMP binding(GO:0016208) |
| 0.2 | 2.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.2 | 0.8 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.2 | 1.9 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.2 | 2.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 1.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.1 | 0.4 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.1 | 1.0 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 2.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.3 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.1 | 1.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.7 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.1 | 1.8 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 2.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.4 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 1.7 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 0.9 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.5 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 4.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 1.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.6 | GO:0051184 | cofactor transporter activity(GO:0051184) |
| 0.1 | 0.7 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.4 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 1.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.5 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 0.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 2.4 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 0.7 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 0.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.7 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 3.4 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.5 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.3 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 1.0 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.3 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 2.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.3 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 1.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 0.5 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 0.4 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 1.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.4 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.6 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 0.5 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 2.0 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.1 | 0.4 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.4 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 4.3 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 1.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 2.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 2.9 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 9.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 6.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.6 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.1 | 0.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.2 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.3 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.1 | 0.6 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 0.3 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 1.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.7 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 0.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 0.2 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 1.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 1.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 1.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 0.4 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 1.1 | GO:0004437 | obsolete inositol or phosphatidylinositol phosphatase activity(GO:0004437) |
| 0.1 | 1.1 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.3 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 5.2 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 0.2 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.1 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 2.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 0.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.4 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.4 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.1 | 2.9 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.1 | 0.8 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.1 | 0.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.2 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 0.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.4 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 0.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.7 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 5.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.4 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 1.8 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.1 | 0.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.5 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.2 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.1 | 0.2 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.1 | 1.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 3.1 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.2 | GO:0015520 | tetracycline:proton antiporter activity(GO:0015520) |
| 0.0 | 2.1 | GO:0032934 | sterol binding(GO:0032934) |
| 0.0 | 1.2 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.8 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.2 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 3.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.6 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.6 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 9.6 | GO:0019787 | ubiquitin-like protein transferase activity(GO:0019787) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.6 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 1.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 1.4 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 3.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 2.8 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.2 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 4.7 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0042978 | prolactin receptor activity(GO:0004925) ornithine decarboxylase activator activity(GO:0042978) ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 1.7 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 2.4 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.5 | GO:0015923 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 18.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.3 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.0 | 4.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 10.2 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 1.7 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.8 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 1.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 14.9 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 1.2 | GO:0050136 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.2 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.2 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 5.1 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.4 | GO:0046873 | metal ion transmembrane transporter activity(GO:0046873) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 2.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.4 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.2 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.1 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 1.6 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 1.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.0 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.8 | GO:0016853 | isomerase activity(GO:0016853) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.0 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 0.1 | GO:0009008 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 4.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 2.0 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 0.1 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.1 | GO:0022829 | wide pore channel activity(GO:0022829) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.4 | SA_TRKA_RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.4 | 3.9 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 6.9 | SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 0.6 | ST_GAQ_PATHWAY | G alpha q Pathway |
| 0.1 | 5.6 | SIG_CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 0.3 | ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | PI3K Pathway |
| 0.1 | 2.4 | SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 1.1 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.7 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.7 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.9 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.7 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.3 | ST_TYPE_I_INTERFERON_PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |