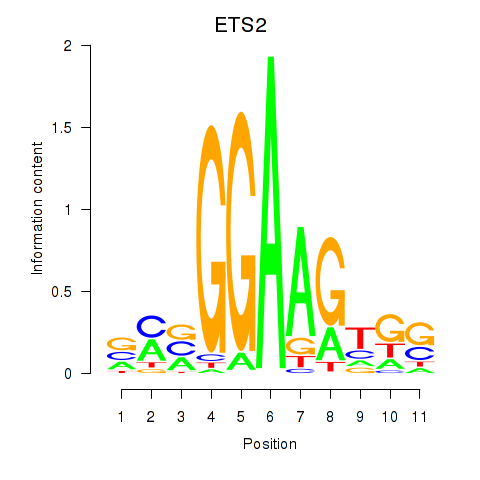
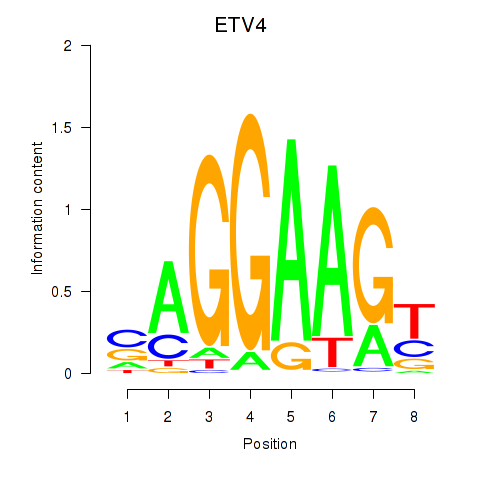
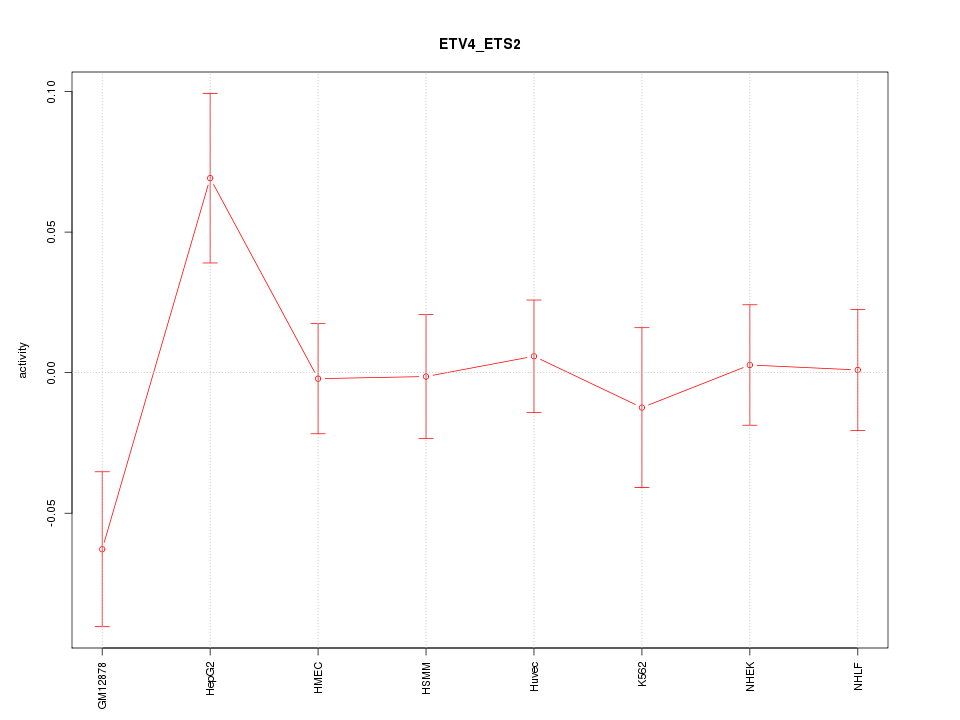
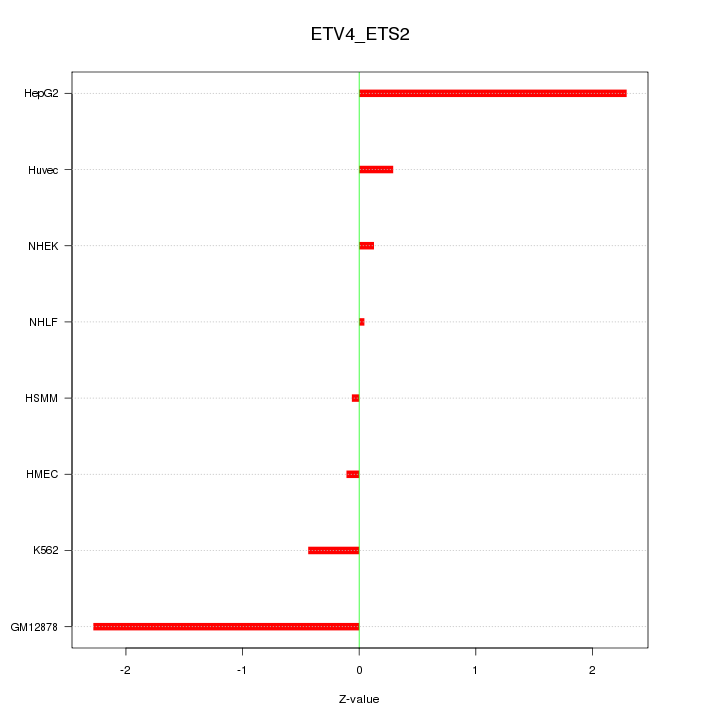
| 1.1 |
4.3 |
GO:0045048 |
protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.9 |
3.5 |
GO:0034442 |
plasma lipoprotein particle oxidation(GO:0034441) regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) lipoprotein modification(GO:0042160) lipoprotein oxidation(GO:0042161) |
| 0.4 |
1.8 |
GO:0060209 |
estrus(GO:0060209) |
| 0.4 |
1.6 |
GO:0001575 |
globoside metabolic process(GO:0001575) |
| 0.3 |
1.0 |
GO:0060729 |
negative regulation of interleukin-23 production(GO:0032707) intestinal epithelial structure maintenance(GO:0060729) |
| 0.3 |
1.2 |
GO:0030037 |
actin filament reorganization involved in cell cycle(GO:0030037) actin filament reorganization(GO:0090527) |
| 0.3 |
4.0 |
GO:0002347 |
response to tumor cell(GO:0002347) |
| 0.3 |
2.3 |
GO:0018126 |
protein hydroxylation(GO:0018126) |
| 0.2 |
0.5 |
GO:0050748 |
negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.2 |
1.1 |
GO:0007614 |
short-term memory(GO:0007614) |
| 0.2 |
1.8 |
GO:0007598 |
blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 |
0.7 |
GO:1904358 |
positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.2 |
1.2 |
GO:2000189 |
regulation of cholesterol homeostasis(GO:2000188) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.2 |
0.8 |
GO:0014067 |
negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.2 |
0.6 |
GO:0070544 |
histone H3-K36 demethylation(GO:0070544) |
| 0.2 |
0.7 |
GO:0006651 |
diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 |
1.1 |
GO:0045945 |
positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.2 |
0.2 |
GO:0002456 |
T cell mediated immunity(GO:0002456) |
| 0.2 |
0.7 |
GO:0060743 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.2 |
0.5 |
GO:0010701 |
positive regulation of chronic inflammatory response(GO:0002678) positive regulation of norepinephrine secretion(GO:0010701) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.2 |
1.6 |
GO:0055003 |
cardiac myofibril assembly(GO:0055003) |
| 0.2 |
2.2 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.2 |
0.5 |
GO:0060318 |
definitive erythrocyte differentiation(GO:0060318) |
| 0.2 |
0.6 |
GO:0055098 |
response to low-density lipoprotein particle(GO:0055098) |
| 0.2 |
2.6 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.2 |
1.1 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.2 |
0.5 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 |
0.4 |
GO:0035269 |
protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.1 |
0.7 |
GO:0032933 |
SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 |
0.9 |
GO:2000172 |
regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.1 |
0.4 |
GO:0002575 |
basophil chemotaxis(GO:0002575) |
| 0.1 |
3.5 |
GO:0043101 |
purine-containing compound salvage(GO:0043101) |
| 0.1 |
0.4 |
GO:0018283 |
metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.1 |
0.1 |
GO:0002467 |
germinal center formation(GO:0002467) |
| 0.1 |
0.8 |
GO:0006011 |
UDP-glucose metabolic process(GO:0006011) UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 |
0.4 |
GO:0003289 |
atrial septum development(GO:0003283) septum primum development(GO:0003284) atrial septum primum morphogenesis(GO:0003289) atrial septum morphogenesis(GO:0060413) |
| 0.1 |
0.3 |
GO:0010792 |
DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 |
0.4 |
GO:0000394 |
RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 |
0.2 |
GO:0030851 |
granulocyte differentiation(GO:0030851) |
| 0.1 |
0.3 |
GO:0043628 |
ncRNA 3'-end processing(GO:0043628) |
| 0.1 |
1.5 |
GO:0030853 |
negative regulation of granulocyte differentiation(GO:0030853) |
| 0.1 |
0.3 |
GO:0034244 |
negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 |
0.8 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.1 |
0.3 |
GO:0071380 |
cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 |
0.3 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.1 |
0.3 |
GO:0060291 |
long-term synaptic potentiation(GO:0060291) |
| 0.1 |
0.3 |
GO:0060586 |
multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.1 |
1.2 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.1 |
1.3 |
GO:0042255 |
ribosome assembly(GO:0042255) |
| 0.1 |
0.6 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.1 |
0.1 |
GO:0032754 |
positive regulation of interleukin-5 production(GO:0032754) |
| 0.1 |
0.2 |
GO:0071504 |
response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.1 |
0.7 |
GO:0019321 |
pentose metabolic process(GO:0019321) |
| 0.1 |
0.2 |
GO:0018243 |
protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 |
0.2 |
GO:1903961 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.1 |
0.4 |
GO:0003197 |
endocardial cushion development(GO:0003197) |
| 0.1 |
0.4 |
GO:0043162 |
ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 |
0.3 |
GO:0050703 |
interleukin-1 alpha secretion(GO:0050703) |
| 0.1 |
0.3 |
GO:0019276 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 |
0.2 |
GO:0043006 |
activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.1 |
0.6 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 |
0.6 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 |
0.4 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 |
0.4 |
GO:0032495 |
response to muramyl dipeptide(GO:0032495) |
| 0.1 |
0.2 |
GO:0032988 |
ribonucleoprotein complex disassembly(GO:0032988) |
| 0.1 |
0.9 |
GO:0034446 |
substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 |
0.2 |
GO:0008356 |
asymmetric cell division(GO:0008356) |
| 0.1 |
0.8 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 |
0.2 |
GO:0033119 |
negative regulation of RNA splicing(GO:0033119) |
| 0.1 |
0.3 |
GO:0008064 |
regulation of actin polymerization or depolymerization(GO:0008064) |
| 0.1 |
0.2 |
GO:0010332 |
response to gamma radiation(GO:0010332) |
| 0.1 |
0.4 |
GO:0042059 |
regulation of epidermal growth factor receptor signaling pathway(GO:0042058) negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) regulation of ERBB signaling pathway(GO:1901184) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.1 |
0.3 |
GO:0090343 |
positive regulation of cell aging(GO:0090343) |
| 0.1 |
0.4 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 |
0.4 |
GO:0060155 |
platelet dense granule organization(GO:0060155) |
| 0.1 |
0.6 |
GO:0042447 |
hormone catabolic process(GO:0042447) |
| 0.1 |
0.6 |
GO:0006998 |
nuclear envelope organization(GO:0006998) |
| 0.1 |
0.2 |
GO:0051083 |
'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 |
0.5 |
GO:0035434 |
copper ion transmembrane transport(GO:0035434) |
| 0.0 |
0.2 |
GO:0045872 |
positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 |
0.2 |
GO:0002903 |
negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 |
0.3 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) |
| 0.0 |
0.2 |
GO:0006434 |
seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 |
0.6 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.0 |
1.4 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.0 |
0.5 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.2 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 |
0.2 |
GO:0015825 |
L-serine transport(GO:0015825) |
| 0.0 |
0.1 |
GO:0010501 |
RNA secondary structure unwinding(GO:0010501) |
| 0.0 |
0.1 |
GO:0042268 |
regulation of cytolysis(GO:0042268) |
| 0.0 |
0.2 |
GO:0003322 |
pancreatic A cell development(GO:0003322) |
| 0.0 |
0.7 |
GO:0048207 |
vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 |
0.3 |
GO:0006515 |
misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 |
0.1 |
GO:0043371 |
negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.0 |
0.5 |
GO:0009650 |
UV protection(GO:0009650) |
| 0.0 |
0.8 |
GO:0006024 |
glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 |
0.4 |
GO:0016998 |
cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 |
0.1 |
GO:0006930 |
substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 |
0.4 |
GO:0072678 |
T cell migration(GO:0072678) |
| 0.0 |
0.5 |
GO:0019228 |
neuronal action potential(GO:0019228) |
| 0.0 |
0.4 |
GO:0000090 |
mitotic anaphase(GO:0000090) |
| 0.0 |
0.1 |
GO:0033278 |
cell proliferation in midbrain(GO:0033278) |
| 0.0 |
0.5 |
GO:0045197 |
establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 |
0.1 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 |
0.5 |
GO:0042274 |
ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 |
1.5 |
GO:0045576 |
mast cell activation(GO:0045576) |
| 0.0 |
0.7 |
GO:0031529 |
ruffle organization(GO:0031529) |
| 0.0 |
0.3 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 |
0.2 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 |
0.1 |
GO:1904396 |
regulation of synaptic growth at neuromuscular junction(GO:0008582) regulation of neuromuscular junction development(GO:1904396) |
| 0.0 |
0.2 |
GO:0007204 |
positive regulation of cytosolic calcium ion concentration(GO:0007204) |
| 0.0 |
0.2 |
GO:0048296 |
isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 |
0.1 |
GO:0010172 |
embryonic body morphogenesis(GO:0010172) |
| 0.0 |
0.2 |
GO:0002667 |
lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 |
0.4 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.6 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.0 |
0.5 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.0 |
0.1 |
GO:0021754 |
facial nucleus development(GO:0021754) |
| 0.0 |
0.5 |
GO:0051881 |
regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 |
0.0 |
GO:0090030 |
regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) regulation of steroid hormone biosynthetic process(GO:0090030) |
| 0.0 |
0.2 |
GO:0051665 |
membrane raft localization(GO:0051665) |
| 0.0 |
0.7 |
GO:0016236 |
macroautophagy(GO:0016236) |
| 0.0 |
0.3 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 |
0.1 |
GO:0010216 |
maintenance of DNA methylation(GO:0010216) |
| 0.0 |
0.0 |
GO:0032621 |
interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.0 |
0.2 |
GO:0006422 |
aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 |
0.3 |
GO:0007512 |
adult heart development(GO:0007512) |
| 0.0 |
0.1 |
GO:0046022 |
regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 |
0.1 |
GO:0002043 |
blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 |
0.2 |
GO:0051974 |
negative regulation of telomerase activity(GO:0051974) |
| 0.0 |
0.5 |
GO:0072595 |
maintenance of protein location in nucleus(GO:0051457) maintenance of protein localization in organelle(GO:0072595) |
| 0.0 |
0.3 |
GO:0045843 |
negative regulation of striated muscle tissue development(GO:0045843) negative regulation of muscle tissue development(GO:1901862) |
| 0.0 |
0.1 |
GO:0061084 |
regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) negative regulation of protein folding(GO:1903333) |
| 0.0 |
0.0 |
GO:0072387 |
flavin adenine dinucleotide metabolic process(GO:0072387) |
| 0.0 |
0.4 |
GO:0043114 |
regulation of vascular permeability(GO:0043114) |
| 0.0 |
0.1 |
GO:0034086 |
maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 |
0.5 |
GO:0000038 |
very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 |
0.3 |
GO:0008634 |
obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.0 |
0.1 |
GO:0021837 |
motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) regulation of mesodermal cell fate specification(GO:0042661) lateral mesoderm development(GO:0048368) lateral mesoderm morphogenesis(GO:0048369) lateral mesoderm formation(GO:0048370) lateral mesodermal cell differentiation(GO:0048371) lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) regulation of mesoderm development(GO:2000380) |
| 0.0 |
0.2 |
GO:0043545 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 |
0.2 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 |
0.0 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 |
0.1 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.1 |
GO:0050983 |
spermidine catabolic process(GO:0046203) deoxyhypusine biosynthetic process from spermidine(GO:0050983) |
| 0.0 |
0.0 |
GO:0060632 |
regulation of microtubule-based movement(GO:0060632) |
| 0.0 |
0.9 |
GO:0030325 |
adrenal gland development(GO:0030325) |
| 0.0 |
0.1 |
GO:0030913 |
paranodal junction assembly(GO:0030913) |
| 0.0 |
0.1 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 |
0.1 |
GO:0031643 |
positive regulation of myelination(GO:0031643) |
| 0.0 |
0.1 |
GO:0034243 |
positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 |
0.1 |
GO:0021527 |
spinal cord association neuron differentiation(GO:0021527) |
| 0.0 |
0.4 |
GO:0003407 |
neural retina development(GO:0003407) |
| 0.0 |
0.1 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.0 |
0.1 |
GO:0048680 |
positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 |
0.2 |
GO:0000183 |
chromatin silencing at rDNA(GO:0000183) |
| 0.0 |
0.1 |
GO:0002032 |
desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 |
0.4 |
GO:0010038 |
response to metal ion(GO:0010038) |
| 0.0 |
0.1 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 |
0.5 |
GO:0018208 |
peptidyl-proline modification(GO:0018208) |
| 0.0 |
0.1 |
GO:0010716 |
regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 |
1.1 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.3 |
GO:1901224 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 |
0.1 |
GO:0014724 |
regulation of twitch skeletal muscle contraction(GO:0014724) fast-twitch skeletal muscle fiber contraction(GO:0031443) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) mitochondrial calcium ion homeostasis(GO:0051560) positive regulation of mitochondrial calcium ion concentration(GO:0051561) relaxation of skeletal muscle(GO:0090076) |
| 0.0 |
0.1 |
GO:1903301 |
regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 |
0.2 |
GO:0006884 |
cell volume homeostasis(GO:0006884) |
| 0.0 |
0.1 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.0 |
0.1 |
GO:0016045 |
detection of bacterium(GO:0016045) |
| 0.0 |
0.1 |
GO:0001824 |
blastocyst development(GO:0001824) |
| 0.0 |
0.0 |
GO:0060054 |
positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 |
0.3 |
GO:0045445 |
myoblast differentiation(GO:0045445) |
| 0.0 |
0.6 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
0.2 |
GO:0051319 |
mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 |
0.1 |
GO:0046475 |
glycerophospholipid catabolic process(GO:0046475) |
| 0.0 |
0.1 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.2 |
GO:0001885 |
endothelial cell development(GO:0001885) |
| 0.0 |
1.2 |
GO:0001889 |
liver development(GO:0001889) hepaticobiliary system development(GO:0061008) |
| 0.0 |
0.2 |
GO:0032922 |
circadian regulation of gene expression(GO:0032922) |
| 0.0 |
1.4 |
GO:0007588 |
excretion(GO:0007588) |
| 0.0 |
0.5 |
GO:0060041 |
retina development in camera-type eye(GO:0060041) |
| 0.0 |
0.3 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 |
1.6 |
GO:0018279 |
peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 |
1.0 |
GO:0007569 |
cell aging(GO:0007569) |
| 0.0 |
0.5 |
GO:0014909 |
smooth muscle cell migration(GO:0014909) |
| 0.0 |
0.2 |
GO:0010821 |
regulation of mitochondrion organization(GO:0010821) |
| 0.0 |
0.2 |
GO:2001021 |
negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.0 |
0.3 |
GO:0045026 |
plasma membrane fusion(GO:0045026) |
| 0.0 |
0.2 |
GO:0006378 |
mRNA polyadenylation(GO:0006378) |
| 0.0 |
0.5 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.0 |
1.5 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.1 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.0 |
0.5 |
GO:0007266 |
Rho protein signal transduction(GO:0007266) |
| 0.0 |
2.6 |
GO:0043087 |
regulation of GTPase activity(GO:0043087) |
| 0.0 |
0.0 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.0 |
0.0 |
GO:0001570 |
vasculogenesis(GO:0001570) |
| 0.0 |
0.3 |
GO:0007176 |
regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 |
0.1 |
GO:0044341 |
sodium-dependent phosphate transport(GO:0044341) |
| 0.0 |
0.0 |
GO:0002384 |
hepatic immune response(GO:0002384) |
| 0.0 |
0.3 |
GO:0030866 |
cortical cytoskeleton organization(GO:0030865) cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 |
0.4 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 |
0.2 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
0.8 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
0.0 |
GO:0046487 |
glyoxylate metabolic process(GO:0046487) |
| 0.0 |
0.3 |
GO:0035338 |
long-chain fatty-acyl-CoA metabolic process(GO:0035336) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 |
0.6 |
GO:0046324 |
regulation of glucose import(GO:0046324) |
| 0.0 |
0.0 |
GO:0033211 |
adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 |
0.1 |
GO:0015695 |
organic cation transport(GO:0015695) |
| 0.0 |
0.2 |
GO:0007171 |
activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 |
0.0 |
GO:0090005 |
negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 |
0.1 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 |
0.2 |
GO:0035036 |
binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 |
0.1 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 |
0.2 |
GO:0051016 |
barbed-end actin filament capping(GO:0051016) |
| 0.0 |
0.2 |
GO:0006004 |
fucose metabolic process(GO:0006004) |
| 0.0 |
1.0 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 |
0.4 |
GO:0007257 |
activation of JUN kinase activity(GO:0007257) |
| 0.0 |
1.6 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.4 |
GO:0097479 |
synaptic vesicle transport(GO:0048489) synaptic vesicle localization(GO:0097479) establishment of synaptic vesicle localization(GO:0097480) |
| 0.0 |
0.0 |
GO:0007341 |
penetration of zona pellucida(GO:0007341) |
| 0.0 |
0.1 |
GO:0006704 |
glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 |
0.2 |
GO:0008038 |
neuron recognition(GO:0008038) |
| 0.0 |
0.2 |
GO:0035050 |
embryonic heart tube development(GO:0035050) |
| 0.0 |
0.0 |
GO:0045065 |
cytotoxic T cell differentiation(GO:0045065) |
| 0.0 |
0.1 |
GO:0001510 |
RNA methylation(GO:0001510) |
| 0.0 |
0.1 |
GO:0071157 |
negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 |
1.0 |
GO:0007160 |
cell-matrix adhesion(GO:0007160) |
| 0.0 |
0.4 |
GO:0000289 |
nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 |
0.1 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.0 |
0.0 |
GO:0002295 |
T-helper cell lineage commitment(GO:0002295) alpha-beta T cell lineage commitment(GO:0002363) CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) regulation of T-helper 17 type immune response(GO:2000316) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 17 cell differentiation(GO:2000319) positive regulation of T-helper 17 cell differentiation(GO:2000321) regulation of T-helper 17 cell lineage commitment(GO:2000328) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 |
0.0 |
GO:0042542 |
response to reactive oxygen species(GO:0000302) response to hydrogen peroxide(GO:0042542) |