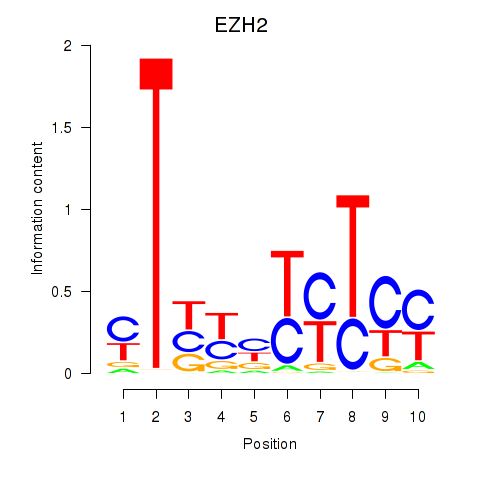
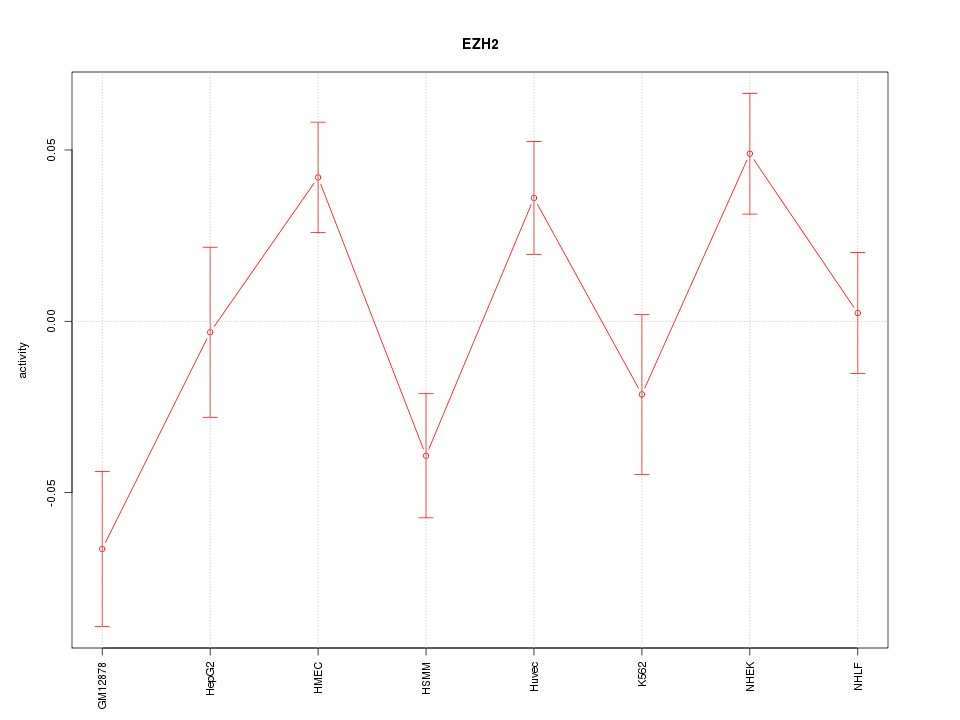
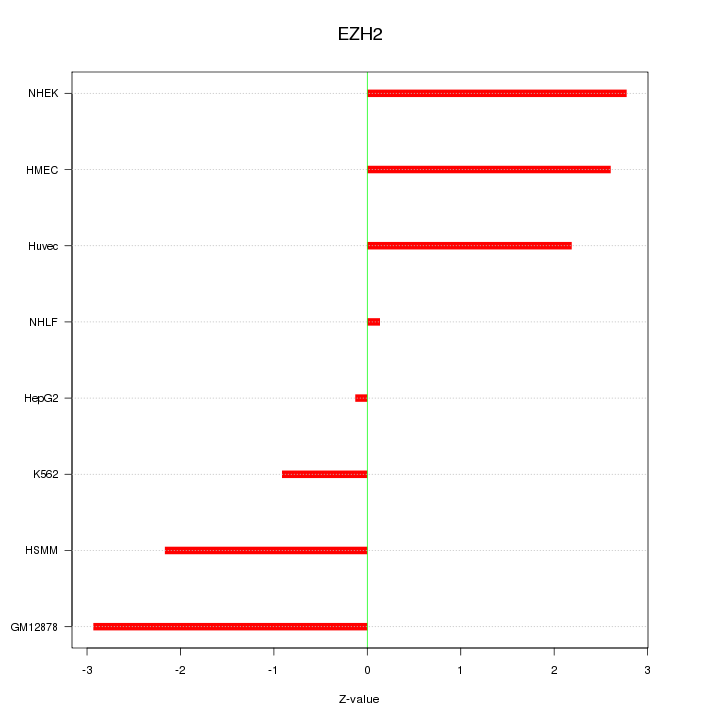
| 1.6 |
6.5 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.7 |
2.0 |
GO:0071639 |
positive regulation of monocyte chemotactic protein-1 production(GO:0071639) |
| 0.6 |
1.7 |
GO:0010982 |
regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.6 |
1.7 |
GO:0048549 |
positive regulation of pinocytosis(GO:0048549) |
| 0.6 |
4.0 |
GO:0090025 |
regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.6 |
2.3 |
GO:0031340 |
positive regulation of vesicle fusion(GO:0031340) |
| 0.5 |
1.9 |
GO:0050928 |
negative regulation of positive chemotaxis(GO:0050928) |
| 0.5 |
2.8 |
GO:2000172 |
regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.5 |
2.8 |
GO:0007499 |
ectoderm and mesoderm interaction(GO:0007499) |
| 0.4 |
1.2 |
GO:0031642 |
negative regulation of myelination(GO:0031642) negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.4 |
1.1 |
GO:0060120 |
auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.3 |
1.2 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.3 |
7.5 |
GO:0045745 |
positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.3 |
3.2 |
GO:0006600 |
creatine metabolic process(GO:0006600) |
| 0.3 |
0.9 |
GO:0060596 |
mammary placode formation(GO:0060596) |
| 0.3 |
1.1 |
GO:0009609 |
response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.3 |
0.8 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.3 |
1.9 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.3 |
0.8 |
GO:0046110 |
xanthine metabolic process(GO:0046110) |
| 0.3 |
0.8 |
GO:0060585 |
regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.3 |
0.8 |
GO:0032488 |
Cdc42 protein signal transduction(GO:0032488) |
| 0.2 |
0.2 |
GO:0031223 |
auditory behavior(GO:0031223) |
| 0.2 |
0.9 |
GO:0009698 |
phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.2 |
0.2 |
GO:0045060 |
negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.2 |
2.8 |
GO:1904893 |
negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 |
1.9 |
GO:0071692 |
sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.2 |
6.0 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.2 |
0.4 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.2 |
1.0 |
GO:0050703 |
interleukin-1 alpha secretion(GO:0050703) |
| 0.2 |
0.8 |
GO:0060649 |
nipple development(GO:0060618) mammary gland bud elongation(GO:0060649) nipple morphogenesis(GO:0060658) nipple sheath formation(GO:0060659) |
| 0.2 |
0.8 |
GO:0048619 |
embryonic hindgut morphogenesis(GO:0048619) |
| 0.2 |
0.6 |
GO:0035283 |
central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.2 |
1.1 |
GO:0043615 |
astrocyte cell migration(GO:0043615) |
| 0.2 |
0.5 |
GO:0048389 |
intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) bud dilation involved in lung branching(GO:0060503) branch elongation involved in ureteric bud branching(GO:0060681) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) pulmonary artery morphogenesis(GO:0061156) cell proliferation involved in mesonephros development(GO:0061209) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) negative regulation of mesonephros development(GO:0061218) pattern specification involved in mesonephros development(GO:0061227) renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) BMP signaling pathway involved in nephric duct formation(GO:0071893) glomerular visceral epithelial cell development(GO:0072015) comma-shaped body morphogenesis(GO:0072049) regulation of branch elongation involved in ureteric bud branching(GO:0072095) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) glomerulus morphogenesis(GO:0072102) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) nephric duct formation(GO:0072179) ureter urothelium development(GO:0072190) ureter epithelial cell differentiation(GO:0072192) ureter morphogenesis(GO:0072197) metanephric comma-shaped body morphogenesis(GO:0072278) glomerular epithelial cell development(GO:0072310) negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.2 |
0.8 |
GO:0090343 |
positive regulation of cell aging(GO:0090343) |
| 0.2 |
0.6 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.2 |
0.5 |
GO:0019056 |
modulation by virus of host transcription(GO:0019056) regulation of sprouting of injured axon(GO:0048686) positive regulation of sprouting of injured axon(GO:0048687) regulation of axon extension involved in regeneration(GO:0048690) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.2 |
0.3 |
GO:0060896 |
neural plate anterior/posterior regionalization(GO:0021999) neural plate pattern specification(GO:0060896) neural plate regionalization(GO:0060897) |
| 0.1 |
0.9 |
GO:0051387 |
negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.1 |
0.4 |
GO:0071635 |
negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.1 |
0.4 |
GO:0046586 |
regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 |
0.4 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.1 |
0.6 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.1 |
0.6 |
GO:0046022 |
regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 |
0.5 |
GO:0045338 |
farnesyl diphosphate metabolic process(GO:0045338) |
| 0.1 |
0.4 |
GO:0060689 |
cell differentiation involved in salivary gland development(GO:0060689) |
| 0.1 |
0.8 |
GO:0046085 |
AMP catabolic process(GO:0006196) adenosine metabolic process(GO:0046085) |
| 0.1 |
0.4 |
GO:0009635 |
response to herbicide(GO:0009635) |
| 0.1 |
3.8 |
GO:0050909 |
sensory perception of taste(GO:0050909) |
| 0.1 |
1.0 |
GO:0045329 |
amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.1 |
0.3 |
GO:0071109 |
superior temporal gyrus development(GO:0071109) |
| 0.1 |
0.8 |
GO:0003065 |
regulation of heart rate by chemical signal(GO:0003062) positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 |
0.3 |
GO:0035269 |
protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.1 |
1.1 |
GO:0008631 |
intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) cell death in response to oxidative stress(GO:0036473) |
| 0.1 |
0.3 |
GO:0001907 |
cytolysis by symbiont of host cells(GO:0001897) killing by symbiont of host cells(GO:0001907) hemolysis by symbiont of host erythrocytes(GO:0019836) disruption by symbiont of host cell(GO:0044004) hemolysis in other organism(GO:0044179) positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) cytolysis in other organism(GO:0051715) cytolysis in other organism involved in symbiotic interaction(GO:0051801) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.1 |
0.9 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 |
0.2 |
GO:0014041 |
regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.1 |
0.7 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 |
1.0 |
GO:0010224 |
response to UV-B(GO:0010224) |
| 0.1 |
1.8 |
GO:0046627 |
negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 |
0.4 |
GO:0010447 |
response to acidic pH(GO:0010447) |
| 0.1 |
0.3 |
GO:0006542 |
glutamine biosynthetic process(GO:0006542) |
| 0.1 |
6.5 |
GO:0006821 |
chloride transport(GO:0006821) |
| 0.1 |
0.4 |
GO:0021615 |
glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 |
0.3 |
GO:0061117 |
cardiac left ventricle formation(GO:0003218) negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.1 |
0.2 |
GO:0048752 |
semicircular canal morphogenesis(GO:0048752) |
| 0.1 |
0.8 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.1 |
0.2 |
GO:1901724 |
positive regulation of cell proliferation involved in kidney development(GO:1901724) |
| 0.1 |
1.2 |
GO:0035307 |
positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 |
0.6 |
GO:0070374 |
regulation of ERK1 and ERK2 cascade(GO:0070372) positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.1 |
0.2 |
GO:0043276 |
anoikis(GO:0043276) |
| 0.1 |
0.9 |
GO:0031529 |
ruffle organization(GO:0031529) |
| 0.1 |
0.4 |
GO:0002019 |
angiotensin catabolic process in blood(GO:0002005) regulation of renal output by angiotensin(GO:0002019) multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) multicellular organismal protein metabolic process(GO:0044268) |
| 0.1 |
1.7 |
GO:0045103 |
intermediate filament-based process(GO:0045103) |
| 0.1 |
0.2 |
GO:0034126 |
regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) positive regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034126) |
| 0.1 |
0.4 |
GO:0032460 |
negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 |
0.2 |
GO:1904037 |
positive regulation of epithelial cell apoptotic process(GO:1904037) positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 |
0.9 |
GO:2001021 |
negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.1 |
1.6 |
GO:0007176 |
regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.1 |
0.6 |
GO:0046051 |
GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.1 |
0.3 |
GO:0043001 |
Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 |
0.2 |
GO:0001575 |
globoside metabolic process(GO:0001575) |
| 0.1 |
1.1 |
GO:0000768 |
syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.1 |
0.2 |
GO:0021978 |
telencephalon regionalization(GO:0021978) |
| 0.1 |
0.2 |
GO:0071918 |
urea transmembrane transport(GO:0071918) |
| 0.1 |
0.1 |
GO:0032071 |
regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.1 |
0.2 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 |
0.2 |
GO:0010157 |
response to chlorate(GO:0010157) |
| 0.1 |
2.4 |
GO:0030520 |
intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 |
0.1 |
GO:0046341 |
CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 |
0.1 |
GO:0033364 |
endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) mast cell secretory granule organization(GO:0033364) |
| 0.0 |
0.2 |
GO:0010919 |
regulation of inositol phosphate biosynthetic process(GO:0010919) positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 |
0.3 |
GO:0016098 |
monoterpenoid metabolic process(GO:0016098) |
| 0.0 |
0.3 |
GO:0048488 |
synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 |
0.2 |
GO:0043922 |
negative regulation by host of viral transcription(GO:0043922) |
| 0.0 |
0.2 |
GO:0006290 |
pyrimidine dimer repair(GO:0006290) |
| 0.0 |
0.1 |
GO:0032897 |
negative regulation of viral transcription(GO:0032897) |
| 0.0 |
0.1 |
GO:0070254 |
mucus secretion(GO:0070254) |
| 0.0 |
1.2 |
GO:0017158 |
regulation of calcium ion-dependent exocytosis(GO:0017158) |
| 0.0 |
0.3 |
GO:0010799 |
regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 |
1.9 |
GO:0090263 |
positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 |
0.5 |
GO:0030949 |
positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 |
0.6 |
GO:0006657 |
CDP-choline pathway(GO:0006657) |
| 0.0 |
0.6 |
GO:0035036 |
binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 |
0.2 |
GO:0030997 |
regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 |
1.2 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 |
1.5 |
GO:0006363 |
termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 |
0.2 |
GO:0001705 |
ectoderm formation(GO:0001705) |
| 0.0 |
0.1 |
GO:0015840 |
urea transport(GO:0015840) |
| 0.0 |
0.5 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.0 |
0.8 |
GO:0032012 |
regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.2 |
GO:0050435 |
beta-amyloid metabolic process(GO:0050435) |
| 0.0 |
0.2 |
GO:0034650 |
aldosterone metabolic process(GO:0032341) aldosterone biosynthetic process(GO:0032342) cortisol metabolic process(GO:0034650) cortisol biosynthetic process(GO:0034651) |
| 0.0 |
0.3 |
GO:0010765 |
positive regulation of sodium ion transport(GO:0010765) |
| 0.0 |
0.3 |
GO:0045920 |
negative regulation of exocytosis(GO:0045920) |
| 0.0 |
0.7 |
GO:0009812 |
flavonoid metabolic process(GO:0009812) |
| 0.0 |
0.5 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
0.3 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.0 |
0.1 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.7 |
GO:0002027 |
regulation of heart rate(GO:0002027) |
| 0.0 |
3.8 |
GO:0006921 |
cellular component disassembly involved in execution phase of apoptosis(GO:0006921) execution phase of apoptosis(GO:0097194) |
| 0.0 |
0.5 |
GO:0051496 |
positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 |
0.5 |
GO:1901224 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 |
0.8 |
GO:0042730 |
fibrinolysis(GO:0042730) |
| 0.0 |
0.0 |
GO:0048520 |
positive regulation of behavior(GO:0048520) |
| 0.0 |
0.4 |
GO:0090083 |
regulation of inclusion body assembly(GO:0090083) |
| 0.0 |
0.3 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 |
3.8 |
GO:0006814 |
sodium ion transport(GO:0006814) |
| 0.0 |
0.1 |
GO:0045872 |
positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 |
0.2 |
GO:0033119 |
negative regulation of RNA splicing(GO:0033119) |
| 0.0 |
0.1 |
GO:0045852 |
pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 |
0.1 |
GO:0021830 |
interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.0 |
0.2 |
GO:0031536 |
positive regulation of exit from mitosis(GO:0031536) |
| 0.0 |
1.6 |
GO:0001764 |
neuron migration(GO:0001764) |
| 0.0 |
0.3 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 |
0.1 |
GO:0007413 |
axonal fasciculation(GO:0007413) |
| 0.0 |
0.1 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.0 |
0.1 |
GO:0006848 |
pyruvate transport(GO:0006848) |
| 0.0 |
0.1 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 |
1.3 |
GO:0071260 |
cellular response to mechanical stimulus(GO:0071260) |
| 0.0 |
0.9 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 |
0.2 |
GO:0022417 |
protein maturation by protein folding(GO:0022417) |
| 0.0 |
0.5 |
GO:0001502 |
cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 |
0.3 |
GO:0007625 |
grooming behavior(GO:0007625) |
| 0.0 |
2.2 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.2 |
GO:0060317 |
cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.0 |
0.0 |
GO:0032291 |
central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 |
0.5 |
GO:0030199 |
collagen fibril organization(GO:0030199) |
| 0.0 |
0.0 |
GO:0048865 |
stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 |
0.1 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 |
0.5 |
GO:0042116 |
macrophage activation(GO:0042116) |
| 0.0 |
0.2 |
GO:0042977 |
activation of JAK2 kinase activity(GO:0042977) |
| 0.0 |
2.9 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.2 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.0 |
0.6 |
GO:0035050 |
embryonic heart tube development(GO:0035050) |
| 0.0 |
0.2 |
GO:0008038 |
neuron recognition(GO:0008038) |
| 0.0 |
0.1 |
GO:0043267 |
negative regulation of potassium ion transport(GO:0043267) |
| 0.0 |
0.2 |
GO:0035162 |
embryonic hemopoiesis(GO:0035162) |
| 0.0 |
0.1 |
GO:0045299 |
otolith mineralization(GO:0045299) |
| 0.0 |
0.1 |
GO:0003351 |
cilium movement(GO:0003341) epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 |
0.1 |
GO:0015910 |
peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 |
0.1 |
GO:0035246 |
peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 |
0.1 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 |
0.2 |
GO:1900047 |
negative regulation of blood coagulation(GO:0030195) negative regulation of wound healing(GO:0061045) negative regulation of hemostasis(GO:1900047) |
| 0.0 |
0.1 |
GO:0043353 |
slow-twitch skeletal muscle fiber contraction(GO:0031444) enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 |
0.1 |
GO:0006540 |
glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 |
0.2 |
GO:0030539 |
male genitalia development(GO:0030539) |
| 0.0 |
0.0 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 |
0.3 |
GO:0030855 |
epithelial cell differentiation(GO:0030855) |
| 0.0 |
0.1 |
GO:0060314 |
regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 |
0.1 |
GO:0003416 |
endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.0 |
0.2 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
0.1 |
GO:0001507 |
acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 |
0.2 |
GO:0045086 |
positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 |
0.1 |
GO:0061081 |
cGMP-mediated signaling(GO:0019934) response to bacterial lipoprotein(GO:0032493) positive regulation of macrophage cytokine production(GO:0060907) positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) response to bacterial lipopeptide(GO:0070339) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) |
| 0.0 |
0.2 |
GO:0070972 |
protein localization to endoplasmic reticulum(GO:0070972) |
| 0.0 |
0.2 |
GO:0046928 |
regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 |
0.2 |
GO:0031214 |
biomineral tissue development(GO:0031214) |
| 0.0 |
0.1 |
GO:0007603 |
phototransduction, visible light(GO:0007603) |
| 0.0 |
0.1 |
GO:0007185 |
transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 |
0.2 |
GO:0051131 |
chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 |
0.1 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 |
0.4 |
GO:0006182 |
cGMP biosynthetic process(GO:0006182) |
| 0.0 |
0.1 |
GO:0051258 |
protein polymerization(GO:0051258) |
| 0.0 |
0.1 |
GO:0009649 |
entrainment of circadian clock(GO:0009649) |
| 0.0 |
0.3 |
GO:0017157 |
regulation of exocytosis(GO:0017157) |
| 0.0 |
0.2 |
GO:0045749 |
obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 |
0.3 |
GO:0018208 |
peptidyl-proline modification(GO:0018208) |
| 0.0 |
0.1 |
GO:0001964 |
startle response(GO:0001964) |
| 0.0 |
0.1 |
GO:0006974 |
cellular response to DNA damage stimulus(GO:0006974) |
| 0.0 |
0.7 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.1 |
GO:0033523 |
histone monoubiquitination(GO:0010390) histone H2B ubiquitination(GO:0033523) |
| 0.0 |
0.3 |
GO:0051101 |
regulation of DNA binding(GO:0051101) |
| 0.0 |
0.2 |
GO:0006953 |
acute-phase response(GO:0006953) |
| 0.0 |
0.3 |
GO:0007566 |
embryo implantation(GO:0007566) |