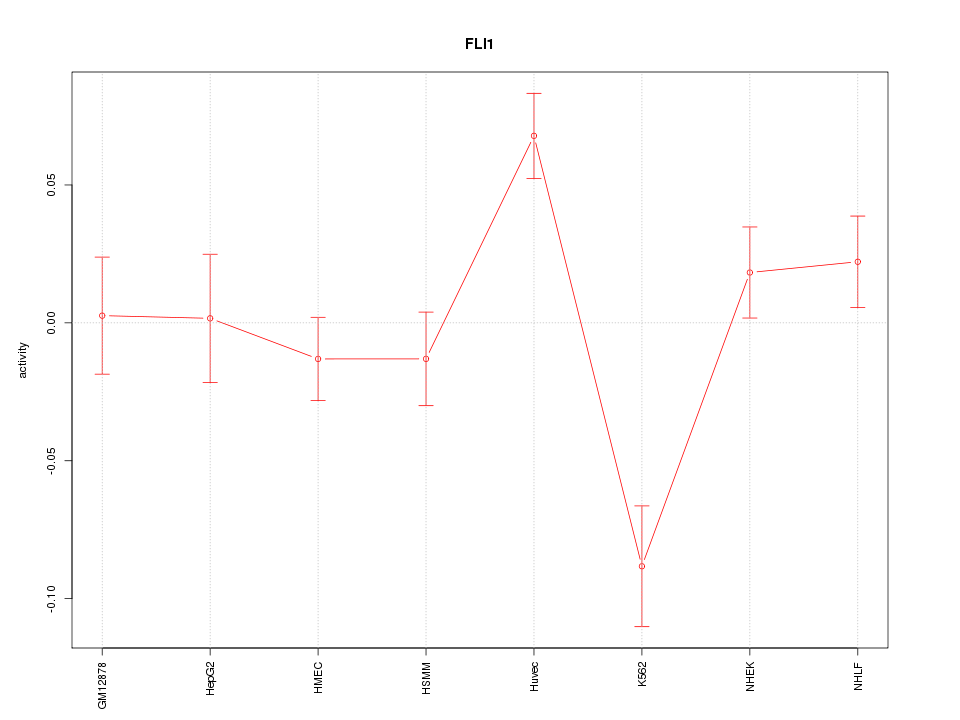
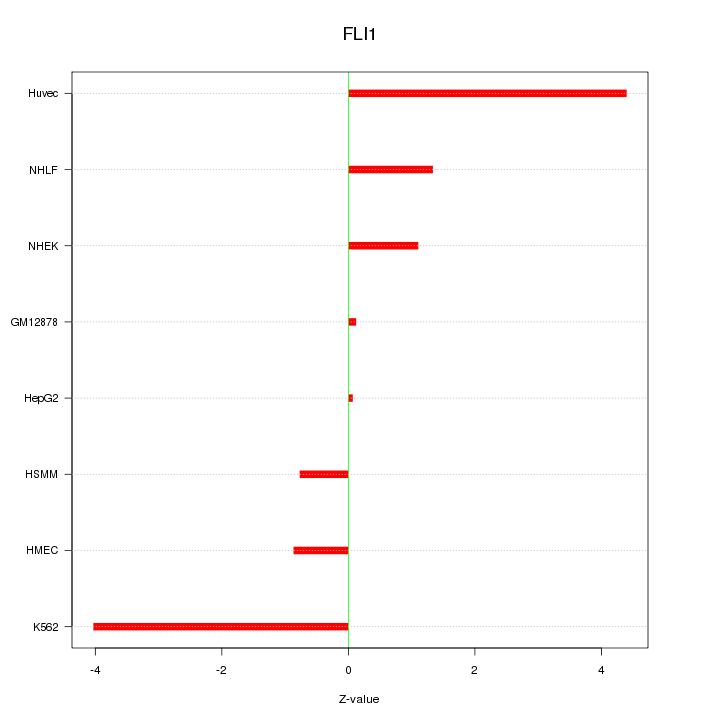
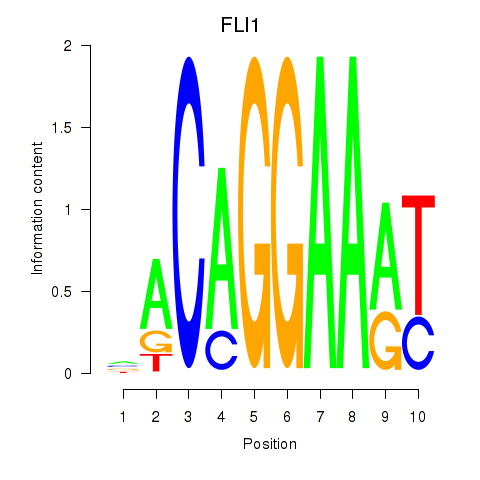
Motif ID: FLI1
Z-value: 2.236
Transcription factors associated with FLI1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| FLI1 | ENSG00000151702.12 | FLI1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.6 | 1.9 | GO:0071380 | cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.6 | 2.5 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.6 | 0.6 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.5 | 1.5 | GO:0043371 | negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.5 | 0.5 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.5 | 1.9 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.5 | 2.3 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.4 | 0.8 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.3 | 1.4 | GO:1904037 | positive regulation of epithelial cell apoptotic process(GO:1904037) positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.3 | 1.7 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.3 | 0.8 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.3 | 2.1 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.2 | 1.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.2 | 1.2 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 | 1.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.2 | 1.0 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 1.4 | GO:0016559 | peroxisome membrane biogenesis(GO:0016557) peroxisome fission(GO:0016559) |
| 0.2 | 0.6 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.2 | 0.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 1.0 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.2 | 1.2 | GO:2000124 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 0.6 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.2 | 0.9 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.2 | 1.8 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.2 | 1.4 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.2 | 0.5 | GO:1903961 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.2 | 1.2 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.2 | 0.5 | GO:0060340 | pericardium morphogenesis(GO:0003344) optic cup formation involved in camera-type eye development(GO:0003408) diencephalon morphogenesis(GO:0048852) positive regulation of type I interferon-mediated signaling pathway(GO:0060340) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.2 | 1.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 | 0.2 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.2 | 1.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.2 | 2.3 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.2 | 1.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 0.6 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.9 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.6 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.7 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.4 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.1 | 0.3 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.1 | 1.1 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 0.8 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 1.6 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.1 | 0.7 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 | 1.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 2.0 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 0.5 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 2.9 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.1 | 1.9 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.1 | 1.0 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 1.1 | GO:0019614 | phenol-containing compound catabolic process(GO:0019336) catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.1 | 0.4 | GO:0034205 | beta-amyloid formation(GO:0034205) |
| 0.1 | 2.9 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.7 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 1.1 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.1 | 0.4 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.1 | 2.2 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.1 | 0.6 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 3.1 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.1 | 0.3 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.1 | 1.8 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.4 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.9 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 4.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 3.1 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 0.9 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.1 | 1.6 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.1 | 2.7 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.1 | 0.3 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 1.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.6 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 1.0 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 0.3 | GO:0034770 | histone H4-K20 methylation(GO:0034770) |
| 0.1 | 1.1 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.1 | 1.3 | GO:0050718 | positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.1 | 0.3 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.1 | 1.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.3 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 2.2 | GO:0042116 | macrophage activation(GO:0042116) |
| 0.1 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.2 | GO:0051892 | negative regulation of heart induction by canonical Wnt signaling pathway(GO:0003136) negative regulation of cell fate specification(GO:0009996) negative regulation of cardioblast cell fate specification(GO:0009997) cardioblast cell fate commitment(GO:0042684) cardioblast cell fate specification(GO:0042685) regulation of cardioblast cell fate specification(GO:0042686) negative regulation of cardioblast differentiation(GO:0051892) cardiac cell fate specification(GO:0060912) canonical Wnt signaling pathway involved in heart development(GO:0061316) negative regulation of heart induction(GO:1901320) negative regulation of cardiocyte differentiation(GO:1905208) regulation of cardiac cell fate specification(GO:2000043) negative regulation of cardiac cell fate specification(GO:2000044) |
| 0.1 | 0.7 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 1.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 1.8 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.1 | 1.8 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.1 | 0.9 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.2 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.1 | 0.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.6 | GO:0006553 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.1 | 0.1 | GO:0032621 | interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.1 | 1.8 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.1 | 0.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.2 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 0.3 | GO:0090042 | tubulin deacetylation(GO:0090042) |
| 0.1 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.1 | 0.4 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 2.8 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.1 | 1.5 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.1 | 0.1 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.1 | 0.3 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.2 | GO:0046520 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.5 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 0.4 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.1 | 0.2 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.1 | 0.2 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 | 0.6 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.1 | 0.4 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.1 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 1.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.3 | GO:0032753 | positive regulation of interleukin-4 production(GO:0032753) |
| 0.1 | 2.6 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 1.0 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.1 | 0.2 | GO:0035269 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.1 | 0.4 | GO:0046851 | negative regulation of odontogenesis(GO:0042483) negative regulation of bone resorption(GO:0045779) negative regulation of bone remodeling(GO:0046851) |
| 0.1 | 0.5 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.1 | 0.6 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.1 | 0.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 1.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.5 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.9 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 1.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) |
| 0.0 | 0.6 | GO:0015669 | gas transport(GO:0015669) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.3 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) |
| 0.0 | 0.7 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.4 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.8 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.7 | GO:1901224 | activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.7 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.2 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.5 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.3 | GO:0006896 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 3.8 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 1.8 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 | 0.1 | GO:2001021 | negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.0 | 3.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) cytoplasmic translation(GO:0002181) cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.9 | GO:0006555 | methionine metabolic process(GO:0006555) |
| 0.0 | 0.4 | GO:0051319 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.1 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.3 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.4 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.5 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 1.0 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.8 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.2 | GO:1901998 | tetracycline transport(GO:0015904) antibiotic transport(GO:0042891) toxin transport(GO:1901998) |
| 0.0 | 1.6 | GO:0045727 | positive regulation of cellular amide metabolic process(GO:0034250) positive regulation of translation(GO:0045727) |
| 0.0 | 0.7 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.4 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.0 | 0.6 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 1.1 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0016045 | detection of bacterium(GO:0016045) |
| 0.0 | 0.0 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.1 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.0 | 0.1 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.4 | GO:0030260 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.1 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.2 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 3.9 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.3 | GO:0018198 | peptidyl-cysteine modification(GO:0018198) |
| 0.0 | 0.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.2 | GO:0044060 | regulation of endocrine process(GO:0044060) |
| 0.0 | 0.3 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0070201 | regulation of establishment of protein localization(GO:0070201) |
| 0.0 | 0.2 | GO:0007389 | pattern specification process(GO:0007389) |
| 0.0 | 0.9 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) |
| 0.0 | 0.2 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.0 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.2 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.3 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.1 | GO:0007204 | positive regulation of cytosolic calcium ion concentration(GO:0007204) regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 0.4 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.1 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.6 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.3 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.4 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.6 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 0.2 | GO:0048265 | response to pain(GO:0048265) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.3 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.3 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) regulation of mesenchymal cell proliferation(GO:0010464) |
| 0.0 | 0.4 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.3 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0048296 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.6 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.6 | GO:0030866 | cortical cytoskeleton organization(GO:0030865) cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.4 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.0 | 0.1 | GO:2000573 | positive regulation of telomerase activity(GO:0051973) positive regulation of DNA biosynthetic process(GO:2000573) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.3 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.0 | 0.4 | GO:0010332 | response to gamma radiation(GO:0010332) |
| 0.0 | 0.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.0 | GO:0046015 | carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.4 | GO:0007632 | visual behavior(GO:0007632) |
| 0.0 | 0.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.1 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.0 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.6 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.2 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.1 | GO:0030030 | cell projection organization(GO:0030030) |
| 0.0 | 0.1 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.2 | GO:0000768 | syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 1.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.5 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.0 | 0.6 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.1 | GO:0045885 | obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.0 | 0.5 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.2 | GO:0033523 | histone monoubiquitination(GO:0010390) histone H2B ubiquitination(GO:0033523) |
| 0.0 | 1.2 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.9 | GO:0043281 | regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043281) |
| 0.0 | 0.9 | GO:0050730 | regulation of peptidyl-tyrosine phosphorylation(GO:0050730) |
| 0.0 | 0.2 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 0.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 1.5 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 3.2 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 1.0 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.0 | GO:0051057 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.6 | 2.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.4 | 2.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.3 | 0.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 0.7 | GO:0032449 | CBM complex(GO:0032449) |
| 0.2 | 0.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 1.0 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) |
| 0.1 | 1.0 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 1.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 4.3 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 3.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.6 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 0.3 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 1.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 0.4 | GO:0014802 | terminal cisterna(GO:0014802) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 1.0 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.1 | 1.0 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 2.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 15.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 0.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.6 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.3 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 1.6 | GO:0019717 | obsolete synaptosome(GO:0019717) |
| 0.1 | 0.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.9 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 5.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 0.3 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.1 | 0.2 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 1.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.8 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.8 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 2.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 2.8 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 5.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.6 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 2.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.4 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.5 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.3 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.0 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 0.6 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.4 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 1.3 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.0 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.1 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 8.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.8 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 1.9 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.7 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.1 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.3 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.9 | GO:0019863 | IgE binding(GO:0019863) |
| 0.6 | 3.9 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.6 | 0.6 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.6 | 1.7 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.6 | 2.8 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.5 | 5.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.4 | 2.2 | GO:0005534 | galactose binding(GO:0005534) |
| 0.4 | 1.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.4 | 2.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.4 | 1.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.4 | 1.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.4 | 1.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.3 | 1.0 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) |
| 0.3 | 2.8 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.3 | 1.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 2.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 1.9 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.2 | 1.4 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 2.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.2 | 1.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.2 | 1.5 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.2 | 1.0 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 1.8 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 1.0 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.2 | 1.9 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.2 | 0.6 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.2 | 1.2 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.2 | 5.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.2 | 1.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.2 | 2.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 0.6 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.2 | 0.6 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 1.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 2.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.4 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 0.7 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 1.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.5 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.2 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 0.5 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 1.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) RNA polymerase II basal transcription factor binding(GO:0001091) TFIIB-class transcription factor binding(GO:0001093) |
| 0.1 | 1.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.5 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.1 | 0.4 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 2.5 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 0.6 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.7 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.7 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.3 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.1 | 1.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.1 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.1 | 0.4 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 2.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.3 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.1 | 0.3 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 0.3 | GO:0008148 | obsolete negative transcription elongation factor activity(GO:0008148) |
| 0.1 | 0.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 1.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.2 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.1 | 0.2 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.1 | 2.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.0 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 1.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.9 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.7 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.3 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.6 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 0.6 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.8 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.2 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 0.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 3.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.2 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.1 | 0.3 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.7 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.1 | 0.3 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 1.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 1.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.9 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.1 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.3 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 1.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 4.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0004769 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.2 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.2 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.6 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.9 | GO:0001104 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 1.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0008493 | tetracycline transporter activity(GO:0008493) |
| 0.0 | 0.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 1.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 1.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.5 | GO:0015645 | fatty acid ligase activity(GO:0015645) |
| 0.0 | 2.6 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 6.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.2 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 1.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.4 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 4.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 2.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 2.5 | GO:0003702 | obsolete RNA polymerase II transcription factor activity(GO:0003702) |
| 0.0 | 1.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 4.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.0 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.2 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.0 | 0.1 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.0 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.4 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.0 | 2.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.0 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0019894 | kinesin binding(GO:0019894) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | ST_IL_13_PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 2.0 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.5 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.6 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.7 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.0 | 0.6 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.3 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |