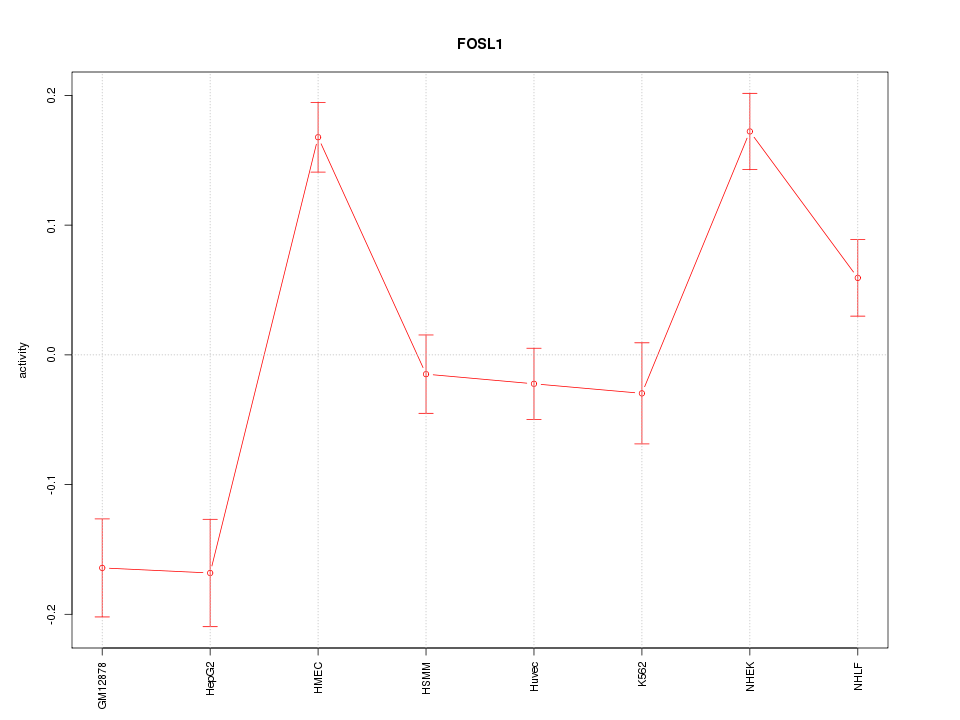
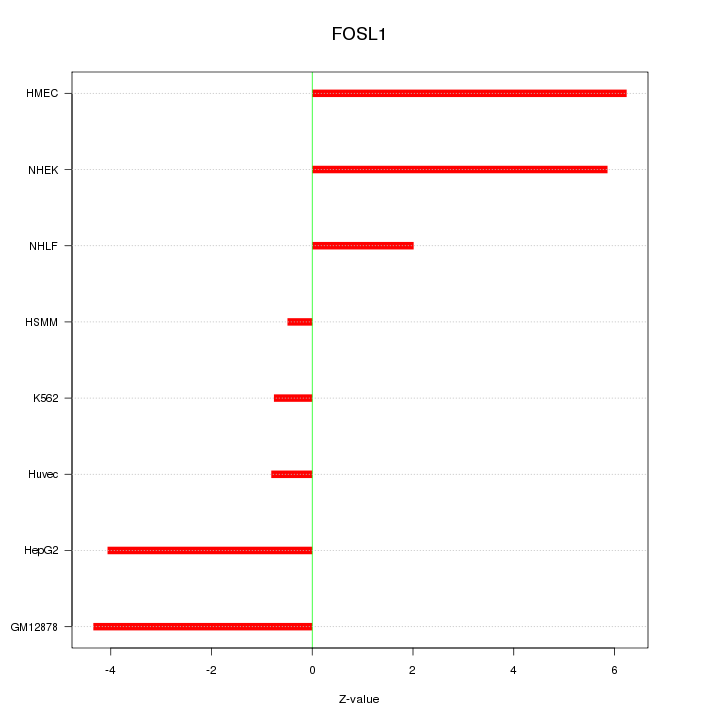
Motif ID: FOSL1
Z-value: 3.779
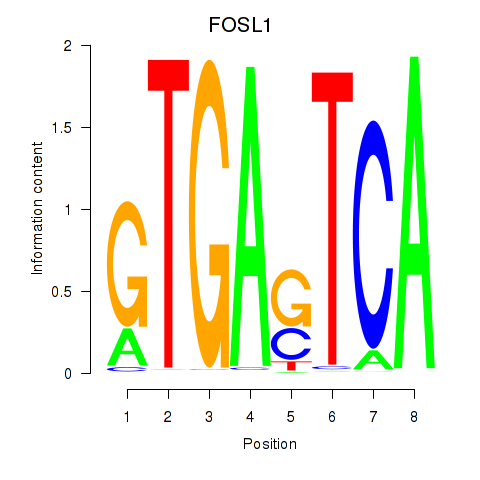
Transcription factors associated with FOSL1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| FOSL1 | ENSG00000175592.4 | FOSL1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 15.9 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 3.4 | 13.5 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 3.3 | 10.0 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 2.6 | 10.3 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 2.5 | 7.4 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 2.4 | 7.3 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 2.4 | 7.3 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 2.3 | 63.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 2.2 | 9.0 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 1.8 | 7.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 1.2 | 4.9 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 1.0 | 9.0 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.9 | 18.9 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.9 | 21.6 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.9 | 4.6 | GO:0090343 | positive regulation of cell aging(GO:0090343) |
| 0.8 | 2.5 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.8 | 25.0 | GO:0031424 | keratinization(GO:0031424) |
| 0.8 | 2.3 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.7 | 9.8 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.7 | 2.7 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.6 | 1.9 | GO:0071603 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) endothelial cell-cell adhesion(GO:0071603) |
| 0.6 | 3.7 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.6 | 3.6 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.6 | 2.9 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.6 | 2.9 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.5 | 1.6 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.5 | 4.7 | GO:0019359 | nicotinamide nucleotide biosynthetic process(GO:0019359) |
| 0.4 | 3.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.4 | 6.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.4 | 9.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.4 | 2.7 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) wound healing, spreading of cells(GO:0044319) epiboly(GO:0090504) epiboly involved in wound healing(GO:0090505) |
| 0.4 | 7.8 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.3 | 8.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.3 | 2.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.3 | 0.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.3 | 1.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.3 | 5.9 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.3 | 1.6 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.3 | 2.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 3.7 | GO:0051927 | obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.2 | 2.0 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.2 | 1.1 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.2 | 1.7 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.2 | 0.5 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.2 | 6.0 | GO:0006309 | DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.2 | 5.4 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.2 | 5.6 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.2 | 2.4 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.2 | 4.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 0.9 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.2 | 22.4 | GO:0008544 | epidermis development(GO:0008544) |
| 0.1 | 0.7 | GO:2000178 | negative regulation of neuroblast proliferation(GO:0007406) response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.1 | 0.4 | GO:0035120 | post-embryonic appendage morphogenesis(GO:0035120) |
| 0.1 | 1.7 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 2.1 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.1 | 5.6 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.1 | 2.7 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.1 | 2.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 4.1 | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) execution phase of apoptosis(GO:0097194) |
| 0.1 | 0.4 | GO:0010759 | complement receptor mediated signaling pathway(GO:0002430) positive regulation of macrophage chemotaxis(GO:0010759) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) positive regulation of neutrophil chemotaxis(GO:0090023) positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 1.8 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.1 | 2.9 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.1 | 0.6 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 0.7 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 0.2 | GO:0043441 | acetoacetic acid biosynthetic process(GO:0043441) |
| 0.1 | 0.3 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.1 | 0.3 | GO:0060100 | regulation of phagocytosis, engulfment(GO:0060099) positive regulation of phagocytosis, engulfment(GO:0060100) regulation of membrane invagination(GO:1905153) positive regulation of membrane invagination(GO:1905155) |
| 0.1 | 3.8 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 2.1 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 4.4 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.8 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.5 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 2.7 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 2.8 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 1.0 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 | 2.9 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.2 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 1.2 | GO:0050931 | melanocyte differentiation(GO:0030318) pigment cell differentiation(GO:0050931) |
| 0.0 | 0.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.3 | GO:0051322 | anaphase(GO:0051322) |
| 0.0 | 0.3 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.3 | GO:1903078 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 | 0.5 | GO:0040013 | negative regulation of cell migration(GO:0030336) negative regulation of locomotion(GO:0040013) negative regulation of cell motility(GO:2000146) |
| 0.0 | 2.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.8 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 1.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.4 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.0 | 1.0 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 | 0.2 | GO:0001960 | negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.0 | 1.5 | GO:0007507 | heart development(GO:0007507) |
| 0.0 | 0.7 | GO:0072164 | ureteric bud development(GO:0001657) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) |
| 0.0 | 0.2 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.0 | 0.2 | GO:0031214 | biomineral tissue development(GO:0031214) |
| 0.0 | 1.8 | GO:0044839 | G2/M transition of mitotic cell cycle(GO:0000086) cell cycle G2/M phase transition(GO:0044839) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 19.0 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 2.7 | 40.3 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 2.2 | 13.3 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 1.4 | 15.9 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 1.2 | 7.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 1.1 | 30.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.7 | 14.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.7 | 2.7 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.5 | 4.6 | GO:0005638 | lamin filament(GO:0005638) |
| 0.5 | 44.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.4 | 1.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.4 | 2.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.3 | 1.7 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.3 | 4.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 2.8 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 2.7 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.2 | 0.7 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.2 | 2.4 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.2 | 1.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.2 | 9.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 1.6 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.1 | 1.9 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.1 | 1.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 2.9 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.1 | 8.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 2.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 1.0 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 6.5 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.1 | 7.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 3.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 22.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 7.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 5.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 2.5 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 1.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 14.3 | GO:0005624 | obsolete membrane fraction(GO:0005624) |
| 0.0 | 1.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 15.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.6 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.1 | GO:0030175 | filopodium(GO:0030175) actin-based cell projection(GO:0098858) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 2.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.4 | GO:0030286 | dynein complex(GO:0030286) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.3 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 1.8 | 7.3 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 1.4 | 8.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 1.4 | 2.7 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 1.2 | 4.7 | GO:0004473 | malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 1.0 | 2.9 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.8 | 7.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.8 | 20.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.7 | 15.9 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.7 | 2.1 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.5 | 6.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.5 | 12.8 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.5 | 8.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.5 | 5.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.5 | 7.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.4 | 12.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.4 | 10.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.4 | 2.3 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.4 | 43.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.4 | 7.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 2.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.3 | 2.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.3 | 14.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.3 | 32.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 1.7 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.3 | 1.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.3 | 7.8 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.2 | 0.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 0.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 3.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 1.1 | GO:0080030 | methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.2 | 0.6 | GO:0050816 | beta-2 adrenergic receptor binding(GO:0031698) phosphothreonine binding(GO:0050816) |
| 0.2 | 3.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 6.0 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.2 | 0.5 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.2 | 3.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 1.8 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 4.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 0.8 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 1.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 2.7 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 1.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 1.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 3.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 18.1 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 9.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.2 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.1 | 1.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 2.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.3 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 9.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 2.0 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 0.2 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.5 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 0.6 | GO:0004835 | tubulin-tyrosine ligase activity(GO:0004835) |
| 0.1 | 0.4 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 4.0 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 39.6 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 0.4 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 0.7 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 1.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 1.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 1.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 1.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.4 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 0.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 6.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 1.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 1.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 19.4 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.9 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 1.6 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 2.4 | GO:0003690 | double-stranded DNA binding(GO:0003690) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 2.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.8 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 5.8 | SA_TRKA_RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.2 | 7.8 | ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | PI3K Pathway |
| 0.2 | 3.7 | SIG_IL4RECEPTOR_IN_B_LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.2 | 7.4 | ST_G_ALPHA_I_PATHWAY | G alpha i Pathway |
| 0.1 | 4.2 | SIG_CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 2.8 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.9 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.9 | SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.2 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |