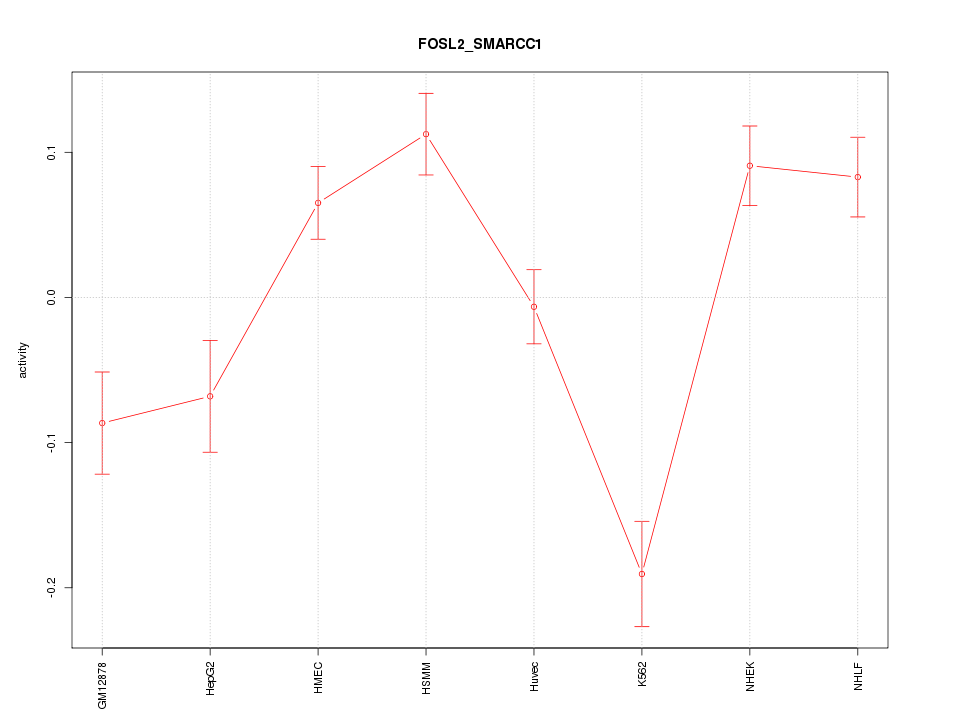
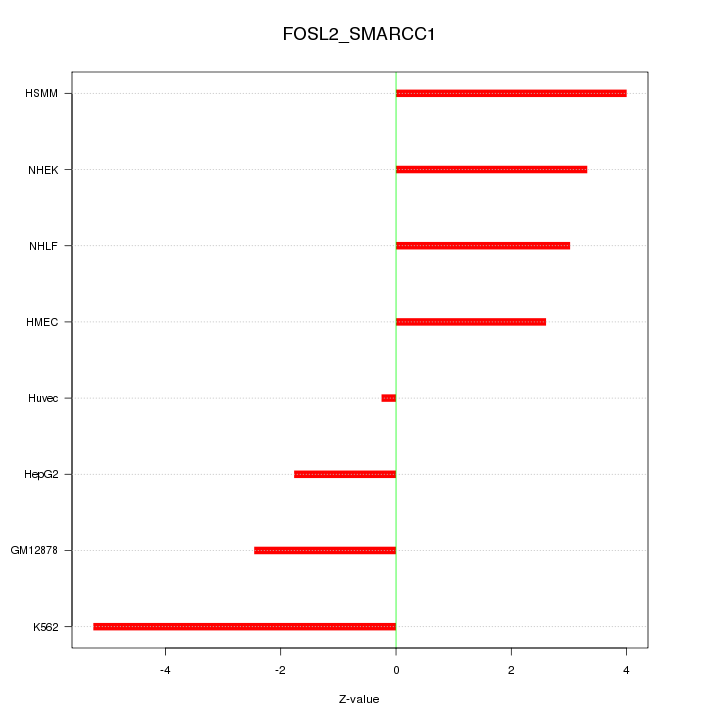
Motif ID: FOSL2_SMARCC1
Z-value: 3.157
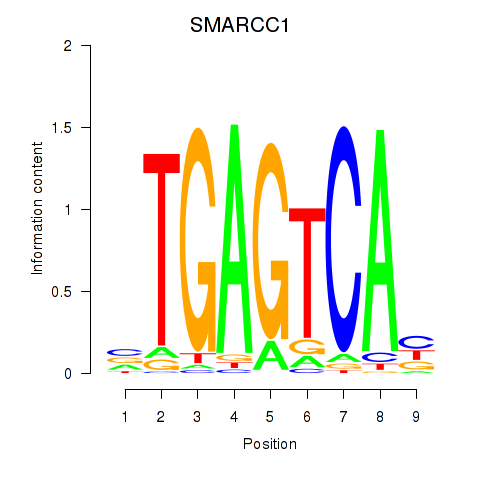
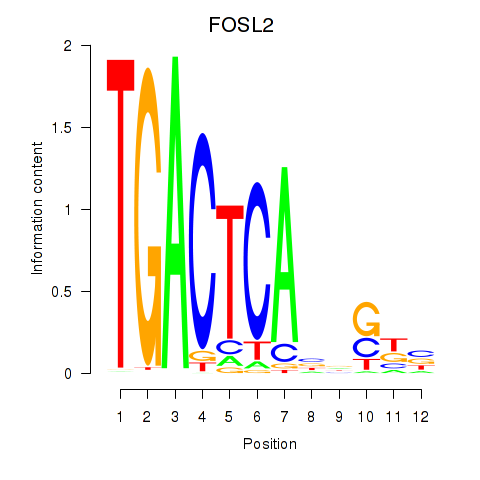
Transcription factors associated with FOSL2_SMARCC1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| FOSL2 | ENSG00000075426.7 | FOSL2 |
| SMARCC1 | ENSG00000173473.6 | SMARCC1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.8 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 1.7 | 6.9 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 1.5 | 4.4 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 1.4 | 41.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 1.2 | 5.0 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 1.2 | 10.7 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 1.1 | 4.5 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 1.1 | 4.4 | GO:0046108 | uridine metabolic process(GO:0046108) |
| 1.1 | 5.5 | GO:0090343 | positive regulation of cell aging(GO:0090343) |
| 1.0 | 7.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.9 | 5.2 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.9 | 3.5 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.8 | 3.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.8 | 3.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.8 | 3.2 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.8 | 5.4 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.8 | 0.8 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.7 | 3.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.7 | 7.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.6 | 2.6 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.6 | 1.9 | GO:0031642 | negative regulation of myelination(GO:0031642) negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.5 | 14.0 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.5 | 2.6 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.5 | 1.5 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.5 | 3.4 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.5 | 0.9 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.5 | 1.4 | GO:0048627 | myoblast development(GO:0048627) |
| 0.5 | 9.2 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.4 | 1.3 | GO:0060547 | negative regulation of necrotic cell death(GO:0060547) |
| 0.4 | 2.9 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.4 | 1.6 | GO:2000178 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.4 | 10.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.3 | 1.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.3 | 1.4 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.3 | 2.4 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.3 | 0.7 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.3 | 2.9 | GO:0019359 | nicotinamide nucleotide biosynthetic process(GO:0019359) |
| 0.3 | 1.6 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 1.9 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.3 | 2.5 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.3 | 1.2 | GO:0017014 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.3 | 0.3 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.3 | 3.6 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.2 | 0.7 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.2 | 1.4 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.2 | 0.9 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.2 | 1.5 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.2 | 2.9 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 | 1.2 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.2 | 2.6 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 | 2.7 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.2 | 0.6 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.2 | 5.7 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.2 | 1.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.2 | 1.5 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.2 | 0.7 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.2 | 0.5 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.2 | 0.9 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 4.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 3.2 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.1 | 0.8 | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) |
| 0.1 | 2.3 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.1 | 5.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 14.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 4.2 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.1 | 5.4 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.1 | 2.1 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.1 | 1.4 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.1 | 0.6 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.5 | GO:0015827 | tryptophan transport(GO:0015827) leucine import(GO:0060356) |
| 0.1 | 2.2 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.1 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.0 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 9.0 | GO:0016573 | histone acetylation(GO:0016573) internal peptidyl-lysine acetylation(GO:0018393) peptidyl-lysine acetylation(GO:0018394) |
| 0.1 | 0.3 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.1 | 0.6 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.1 | 4.4 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 1.6 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.2 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.1 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 2.7 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.1 | 1.1 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.1 | 0.2 | GO:0060291 | long-term synaptic potentiation(GO:0060291) long term synaptic depression(GO:0060292) |
| 0.1 | 9.4 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 0.2 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 | 0.1 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.1 | 1.0 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 2.1 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.1 | 0.1 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.1 | 0.9 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 1.7 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 0.9 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 1.8 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 4.2 | GO:0009408 | response to heat(GO:0009408) |
| 0.1 | 1.1 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 0.2 | GO:0070942 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) |
| 0.1 | 8.6 | GO:0008544 | epidermis development(GO:0008544) |
| 0.1 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0021837 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) regulation of mesodermal cell fate specification(GO:0042661) lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) regulation of mesoderm development(GO:2000380) |
| 0.0 | 1.2 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.4 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.2 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 2.2 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 5.3 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.2 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 3.4 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.0 | 1.2 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.0 | 2.3 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.3 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.9 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 1.1 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 4.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.5 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 1.2 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.4 | GO:0051271 | negative regulation of cell migration(GO:0030336) negative regulation of locomotion(GO:0040013) negative regulation of cellular component movement(GO:0051271) negative regulation of cell motility(GO:2000146) |
| 0.0 | 0.4 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 2.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 6.3 | GO:0016049 | cell growth(GO:0016049) |
| 0.0 | 0.3 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.3 | GO:0045047 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 1.8 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.0 | 3.9 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.2 | GO:0030260 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.5 | GO:0051925 | obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.0 | 2.0 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 1.1 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.0 | 0.2 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.7 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 2.4 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.1 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.3 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 0.7 | GO:0051291 | protein heterooligomerization(GO:0051291) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 33.6 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 1.9 | 1.9 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 1.1 | 6.8 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 1.1 | 6.5 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 1.1 | 2.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.9 | 3.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.9 | 4.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.7 | 3.4 | GO:0032449 | CBM complex(GO:0032449) |
| 0.7 | 2.0 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.6 | 5.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.6 | 2.4 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.6 | 2.8 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.6 | 7.8 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.4 | 2.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.4 | 2.5 | GO:1990752 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.3 | 2.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.3 | 3.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 3.6 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.2 | 2.2 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.2 | 0.6 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.2 | 17.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 2.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 7.5 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 1.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 6.1 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 0.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 2.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 5.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 1.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 3.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 5.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 2.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 30.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 12.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 4.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 3.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.1 | 0.9 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.1 | 5.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 2.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 1.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 3.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.0 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 1.2 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 2.6 | GO:0044309 | neuron spine(GO:0044309) |
| 0.1 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 6.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 2.3 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 1.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 2.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.4 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 5.8 | GO:0005856 | cytoskeleton(GO:0005856) |
| 0.0 | 2.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.8 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.7 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 13.0 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 2.3 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 19.2 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.1 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 1.7 | 6.8 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 1.2 | 7.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 1.1 | 4.4 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.9 | 5.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.8 | 2.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.7 | 2.9 | GO:0004473 | malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.7 | 2.2 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.6 | 11.0 | GO:0043236 | laminin binding(GO:0043236) |
| 0.6 | 2.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.6 | 14.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.5 | 13.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.5 | 1.5 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.5 | 4.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.4 | 2.0 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.4 | 4.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.4 | 9.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 2.7 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.3 | 1.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.3 | 2.6 | GO:0051425 | acetylcholine receptor binding(GO:0033130) PTB domain binding(GO:0051425) |
| 0.3 | 6.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.3 | 13.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.3 | 1.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.3 | 5.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.3 | 0.8 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.2 | 4.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 0.7 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.2 | 3.0 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.2 | 1.2 | GO:0032558 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.2 | 0.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 0.9 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 1.6 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 2.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 2.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 1.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 17.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 4.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 0.7 | GO:0000739 | obsolete DNA strand annealing activity(GO:0000739) |
| 0.2 | 4.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 1.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 15.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 4.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 3.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 2.8 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 13.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.6 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 1.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 1.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 4.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.6 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 2.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.7 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.3 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.1 | 1.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.6 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 2.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.0 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.8 | GO:0004835 | tubulin-tyrosine ligase activity(GO:0004835) |
| 0.1 | 2.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 5.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 6.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 5.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.2 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 0.2 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.1 | 6.7 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 0.2 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.2 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.1 | 1.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 0.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 4.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 0.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 34.6 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 1.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 9.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.9 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.6 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 1.4 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.0 | 29.4 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0015368 | calcium:cation antiporter activity(GO:0015368) |
| 0.0 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 2.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 1.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 6.0 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.2 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.0 | 0.4 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 1.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 2.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) signaling adaptor activity(GO:0035591) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.5 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 1.8 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 3.1 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.1 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.0 | 1.0 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 2.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.7 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 5.1 | ST_G_ALPHA_I_PATHWAY | G alpha i Pathway |
| 0.1 | 5.5 | SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 2.8 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.7 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.4 | ST_STAT3_PATHWAY | STAT3 Pathway |
| 0.0 | 1.1 | SIG_BCR_SIGNALING_PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.7 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.3 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |