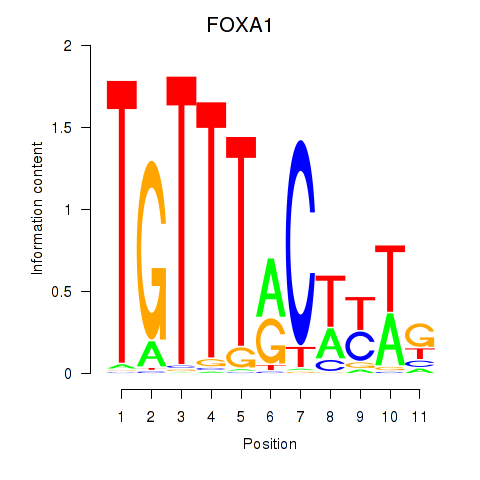
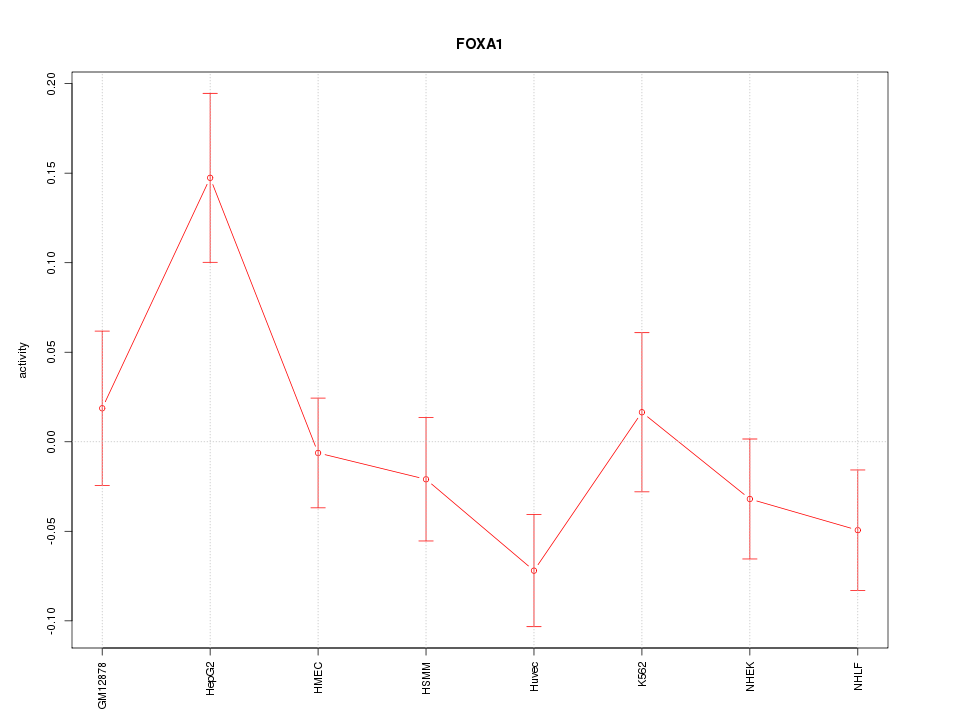
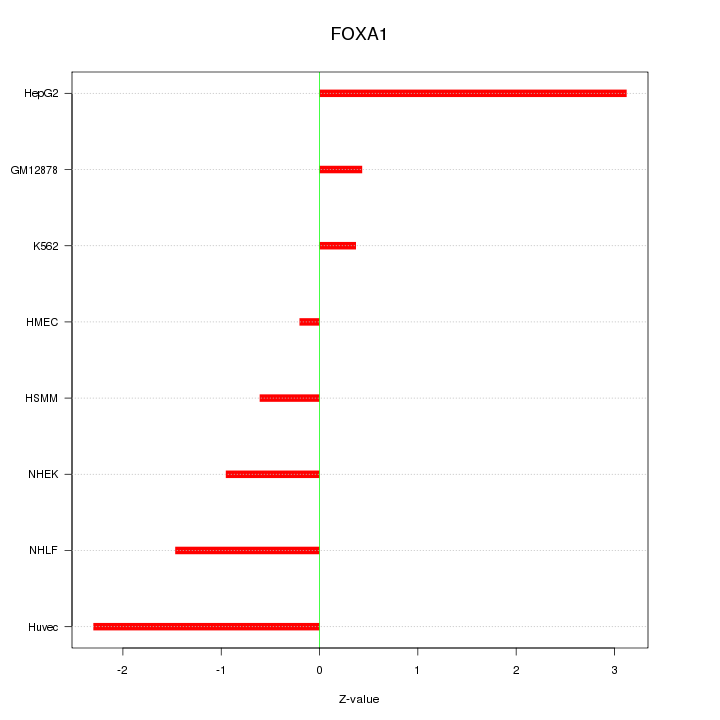
| 1.6 |
6.5 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 1.2 |
4.9 |
GO:0071379 |
cellular response to prostaglandin stimulus(GO:0071379) |
| 0.7 |
4.6 |
GO:0045780 |
positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.4 |
1.3 |
GO:0046521 |
sphingoid catabolic process(GO:0046521) |
| 0.4 |
1.6 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.4 |
1.2 |
GO:0055005 |
optic placode formation involved in camera-type eye formation(GO:0046619) ventricular cardiac myofibril assembly(GO:0055005) embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.3 |
1.0 |
GO:0035623 |
renal glucose absorption(GO:0035623) |
| 0.3 |
1.9 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.2 |
1.1 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 |
1.0 |
GO:0045953 |
desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) negative regulation of natural killer cell mediated immunity(GO:0002716) negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.2 |
1.3 |
GO:0060272 |
embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.2 |
3.9 |
GO:0006825 |
copper ion transport(GO:0006825) |
| 0.2 |
0.5 |
GO:0045835 |
negative regulation of meiotic nuclear division(GO:0045835) |
| 0.2 |
4.0 |
GO:0008211 |
glucocorticoid metabolic process(GO:0008211) |
| 0.2 |
2.2 |
GO:0060706 |
cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.1 |
0.4 |
GO:0000114 |
obsolete regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0000114) |
| 0.1 |
0.5 |
GO:0051919 |
positive regulation of fibrinolysis(GO:0051919) |
| 0.1 |
0.3 |
GO:0007518 |
myoblast fate determination(GO:0007518) |
| 0.1 |
0.4 |
GO:1904037 |
positive regulation of epithelial cell apoptotic process(GO:1904037) positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 |
0.3 |
GO:0050917 |
sensory perception of umami taste(GO:0050917) |
| 0.1 |
0.3 |
GO:0001544 |
initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 |
0.3 |
GO:0010430 |
ethanol catabolic process(GO:0006068) fatty acid omega-oxidation(GO:0010430) |
| 0.1 |
4.3 |
GO:0046324 |
regulation of glucose import(GO:0046324) |
| 0.1 |
0.2 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.1 |
0.3 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) |
| 0.1 |
1.5 |
GO:0008210 |
estrogen metabolic process(GO:0008210) |
| 0.1 |
0.3 |
GO:0010157 |
response to chlorate(GO:0010157) |
| 0.1 |
0.2 |
GO:0032416 |
negative regulation of sodium:proton antiporter activity(GO:0032416) negative regulation of sodium ion transmembrane transport(GO:1902306) negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 |
1.4 |
GO:0045165 |
cell fate commitment(GO:0045165) |
| 0.1 |
2.9 |
GO:0031100 |
organ regeneration(GO:0031100) |
| 0.1 |
0.2 |
GO:0035987 |
endodermal cell fate commitment(GO:0001711) endodermal cell differentiation(GO:0035987) |
| 0.1 |
1.8 |
GO:1900047 |
negative regulation of blood coagulation(GO:0030195) negative regulation of wound healing(GO:0061045) negative regulation of hemostasis(GO:1900047) |
| 0.0 |
0.2 |
GO:2000831 |
renin-angiotensin regulation of aldosterone production(GO:0002018) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) positive regulation of NAD(P)H oxidase activity(GO:0033864) corticosteroid hormone secretion(GO:0035930) mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of steroid hormone secretion(GO:2000831) regulation of corticosteroid hormone secretion(GO:2000846) regulation of mineralocorticoid secretion(GO:2000855) regulation of aldosterone secretion(GO:2000858) |
| 0.0 |
0.4 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.0 |
0.9 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.0 |
0.2 |
GO:0090231 |
regulation of spindle checkpoint(GO:0090231) |
| 0.0 |
0.8 |
GO:0045730 |
respiratory burst(GO:0045730) |
| 0.0 |
1.6 |
GO:0043154 |
negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.0 |
0.2 |
GO:0060259 |
regulation of feeding behavior(GO:0060259) |
| 0.0 |
0.4 |
GO:0031641 |
regulation of myelination(GO:0031641) |
| 0.0 |
0.1 |
GO:0042759 |
fatty acid elongation, saturated fatty acid(GO:0019367) long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 |
2.9 |
GO:0010951 |
negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 |
0.5 |
GO:0006657 |
CDP-choline pathway(GO:0006657) |
| 0.0 |
1.1 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.0 |
0.5 |
GO:0046627 |
negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 |
0.3 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.0 |
0.9 |
GO:0006024 |
glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 |
0.7 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 |
0.3 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 |
0.1 |
GO:0050747 |
positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 |
0.4 |
GO:0001893 |
maternal placenta development(GO:0001893) |
| 0.0 |
0.1 |
GO:0045084 |
positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 |
0.2 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.2 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.0 |
0.1 |
GO:0046621 |
negative regulation of organ growth(GO:0046621) |
| 0.0 |
0.1 |
GO:0046642 |
negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 |
1.8 |
GO:0007586 |
digestion(GO:0007586) |
| 0.0 |
0.5 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.1 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 |
1.6 |
GO:0048015 |
phosphatidylinositol-mediated signaling(GO:0048015) inositol lipid-mediated signaling(GO:0048017) |
| 0.0 |
0.1 |
GO:1900087 |
traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.0 |
0.1 |
GO:0045085 |
negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 |
0.1 |
GO:0060346 |
bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.0 |
0.7 |
GO:0007416 |
synapse assembly(GO:0007416) |
| 0.0 |
1.7 |
GO:0007179 |
transforming growth factor beta receptor signaling pathway(GO:0007179) |