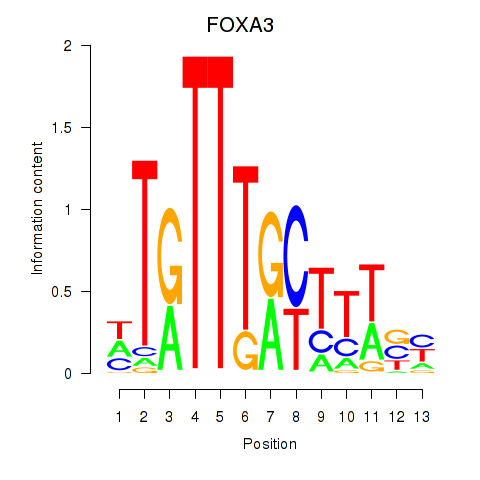
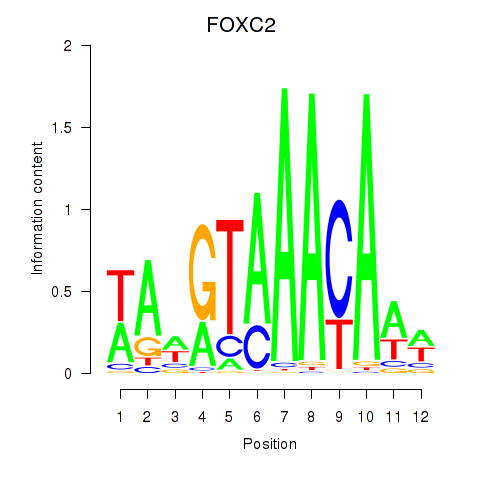
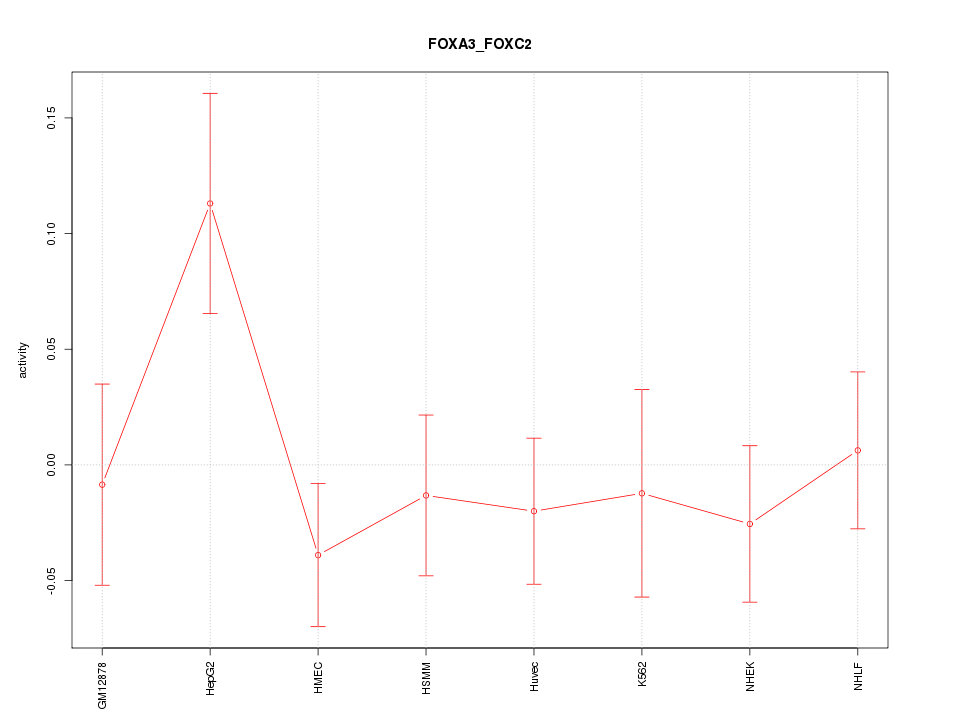
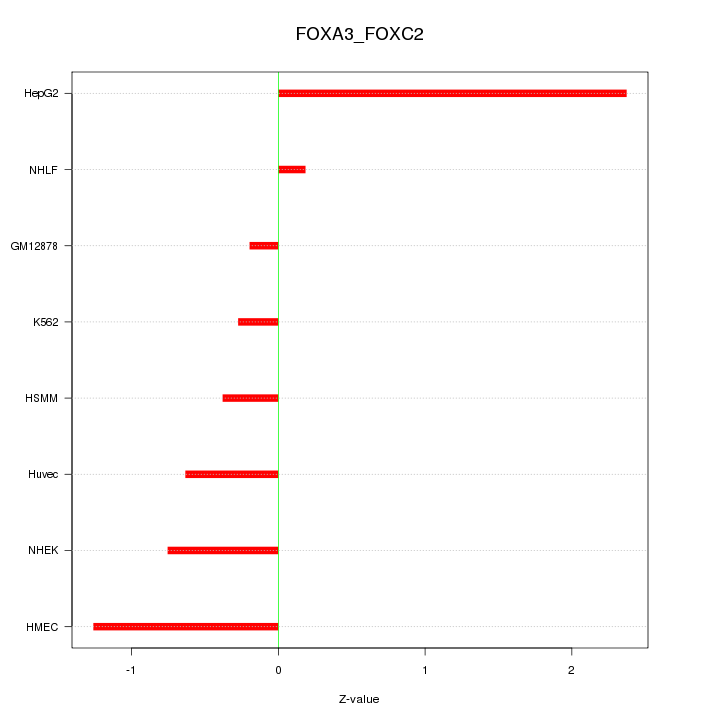
| 1.1 |
4.4 |
GO:0010903 |
negative regulation of cytokine secretion involved in immune response(GO:0002740) negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 1.1 |
4.3 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.6 |
1.9 |
GO:0001907 |
cytolysis by symbiont of host cells(GO:0001897) killing by symbiont of host cells(GO:0001907) hemolysis by symbiont of host erythrocytes(GO:0019836) disruption by symbiont of host cell(GO:0044004) hemolysis in other organism(GO:0044179) positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) cytolysis in other organism(GO:0051715) cytolysis in other organism involved in symbiotic interaction(GO:0051801) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.6 |
2.4 |
GO:0071379 |
cellular response to prostaglandin stimulus(GO:0071379) |
| 0.4 |
2.0 |
GO:0006651 |
diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 |
0.6 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.2 |
0.8 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 |
0.7 |
GO:0051919 |
positive regulation of fibrinolysis(GO:0051919) |
| 0.2 |
0.7 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.2 |
0.8 |
GO:0045953 |
desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) negative regulation of natural killer cell mediated immunity(GO:0002716) negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.2 |
2.5 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 |
0.4 |
GO:0007518 |
myoblast fate determination(GO:0007518) |
| 0.1 |
0.3 |
GO:0046521 |
sphingoid catabolic process(GO:0046521) |
| 0.1 |
1.1 |
GO:0031641 |
regulation of myelination(GO:0031641) |
| 0.1 |
2.3 |
GO:0031000 |
response to caffeine(GO:0031000) |
| 0.1 |
0.2 |
GO:0042320 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) |
| 0.1 |
1.0 |
GO:0060706 |
cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.1 |
0.7 |
GO:0006069 |
ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.1 |
0.6 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.1 |
1.7 |
GO:0008211 |
glucocorticoid metabolic process(GO:0008211) |
| 0.1 |
4.1 |
GO:0046324 |
regulation of glucose import(GO:0046324) |
| 0.1 |
0.5 |
GO:0030277 |
epithelial structure maintenance(GO:0010669) maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 |
0.2 |
GO:0015825 |
L-serine transport(GO:0015825) |
| 0.0 |
0.2 |
GO:0046642 |
negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 |
0.7 |
GO:0034446 |
substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 |
1.0 |
GO:0008210 |
estrogen metabolic process(GO:0008210) |
| 0.0 |
0.1 |
GO:0052257 |
pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 |
0.2 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 |
0.8 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.0 |
0.1 |
GO:0000114 |
obsolete regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0000114) |
| 0.0 |
0.1 |
GO:0001544 |
initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 |
0.1 |
GO:0021527 |
spinal cord association neuron differentiation(GO:0021527) |
| 0.0 |
3.1 |
GO:0007586 |
digestion(GO:0007586) |
| 0.0 |
0.5 |
GO:0000038 |
very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 |
0.1 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.0 |
0.1 |
GO:0032416 |
negative regulation of sodium:proton antiporter activity(GO:0032416) negative regulation of sodium ion transmembrane transport(GO:1902306) negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 |
0.1 |
GO:0032410 |
negative regulation of transporter activity(GO:0032410) |
| 0.0 |
0.3 |
GO:0046627 |
negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 |
0.6 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.0 |
0.2 |
GO:0007185 |
transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 |
0.2 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 |
0.1 |
GO:0007492 |
endoderm development(GO:0007492) |
| 0.0 |
0.1 |
GO:0006553 |
lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 |
0.6 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.0 |
0.1 |
GO:2000831 |
renin-angiotensin regulation of aldosterone production(GO:0002018) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) positive regulation of NAD(P)H oxidase activity(GO:0033864) corticosteroid hormone secretion(GO:0035930) mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of steroid hormone secretion(GO:2000831) regulation of corticosteroid hormone secretion(GO:2000846) regulation of mineralocorticoid secretion(GO:2000855) regulation of aldosterone secretion(GO:2000858) |
| 0.0 |
0.1 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.0 |
0.6 |
GO:0043154 |
negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 |
0.5 |
GO:0006024 |
glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 |
0.1 |
GO:0044341 |
sodium-dependent phosphate transport(GO:0044341) |
| 0.0 |
0.6 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
1.5 |
GO:0002576 |
platelet degranulation(GO:0002576) |
| 0.0 |
1.1 |
GO:0010951 |
negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 |
0.1 |
GO:0016576 |
histone dephosphorylation(GO:0016576) |
| 0.0 |
0.2 |
GO:0001960 |
negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.0 |
0.1 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 |
0.2 |
GO:0045744 |
negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 |
0.1 |
GO:0008272 |
sulfate transport(GO:0008272) |
| 0.0 |
0.3 |
GO:0032570 |
response to progesterone(GO:0032570) |
| 0.0 |
1.0 |
GO:0007605 |
sensory perception of sound(GO:0007605) |