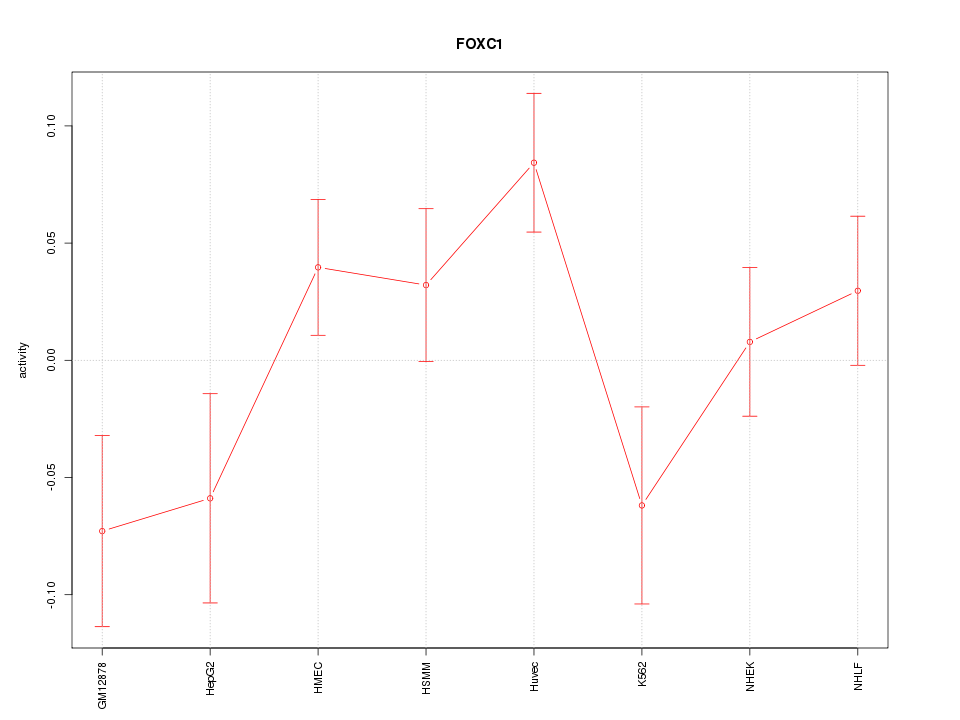
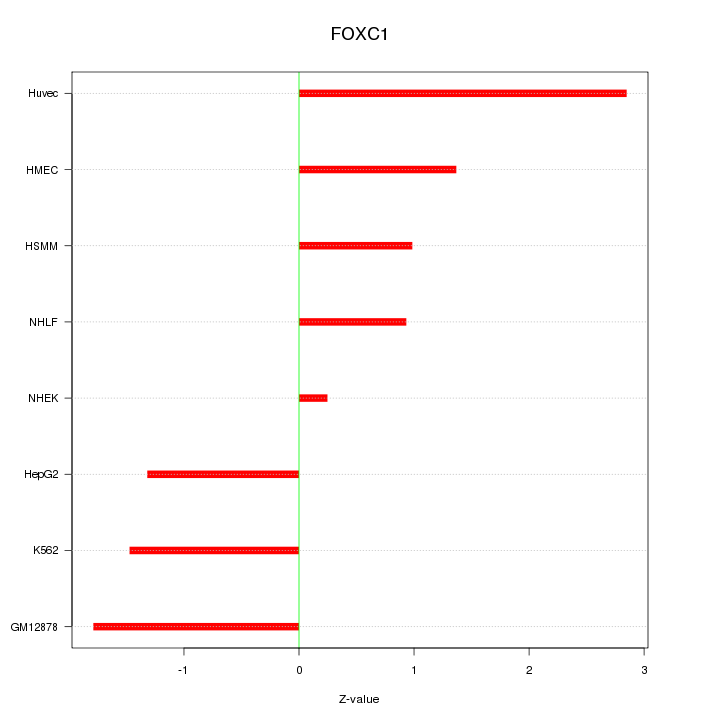
Motif ID: FOXC1
Z-value: 1.541
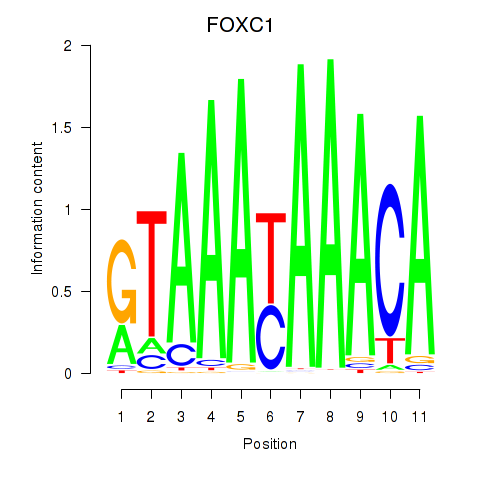
Transcription factors associated with FOXC1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| FOXC1 | ENSG00000054598.5 | FOXC1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.0 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 1.1 | 3.4 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.4 | 3.2 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.3 | 1.4 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.3 | 1.0 | GO:0050758 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.3 | 1.9 | GO:2000124 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 1.2 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.2 | 1.5 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.2 | 0.7 | GO:0071635 | negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.2 | 2.3 | GO:0010658 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.2 | 1.6 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.2 | 0.6 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.2 | 5.6 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.2 | 0.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.6 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.9 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 2.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.9 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.1 | 0.3 | GO:0043276 | anoikis(GO:0043276) |
| 0.1 | 0.3 | GO:0003171 | atrioventricular valve development(GO:0003171) aortic valve development(GO:0003176) aortic valve morphogenesis(GO:0003180) atrioventricular valve morphogenesis(GO:0003181) notochord formation(GO:0014028) notochord morphogenesis(GO:0048570) |
| 0.1 | 0.5 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 0.8 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.2 | GO:0070781 | response to biotin(GO:0070781) |
| 0.1 | 0.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.4 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.2 | GO:0050653 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 0.3 | GO:0045080 | positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.1 | 2.5 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) |
| 0.0 | 0.4 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:0015811 | sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.0 | 0.6 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.1 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.3 | GO:0045329 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 | 1.5 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.6 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.1 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 1.4 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 1.1 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 1.9 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.5 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 3.3 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.8 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.0 | GO:0007285 | primary spermatocyte growth(GO:0007285) |
| 0.0 | 0.2 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0043030 | regulation of macrophage activation(GO:0043030) |
| 0.0 | 0.3 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.0 | 0.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.9 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 1.3 | 5.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.8 | 2.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 9.0 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 0.6 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.8 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.3 | GO:0019718 | obsolete rough microsome(GO:0019718) |
| 0.1 | 1.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.8 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.5 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.6 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.3 | 3.4 | GO:0030547 | receptor inhibitor activity(GO:0030547) receptor antagonist activity(GO:0048019) |
| 0.3 | 3.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.2 | 1.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 2.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.2 | 1.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.2 | 2.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) RNA polymerase II basal transcription factor binding(GO:0001091) TFIIB-class transcription factor binding(GO:0001093) |
| 0.1 | 1.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.6 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 1.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.9 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 1.6 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 3.3 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.2 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.1 | 0.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 8.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.2 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.1 | 1.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 0.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 1.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 1.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 3.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 8.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.2 | GO:0046934 | obsolete inositol or phosphatidylinositol kinase activity(GO:0004428) phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.6 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 2.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 5.4 | SIG_BCR_SIGNALING_PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 1.0 | ST_G_ALPHA_I_PATHWAY | G alpha i Pathway |
| 0.0 | 0.9 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |