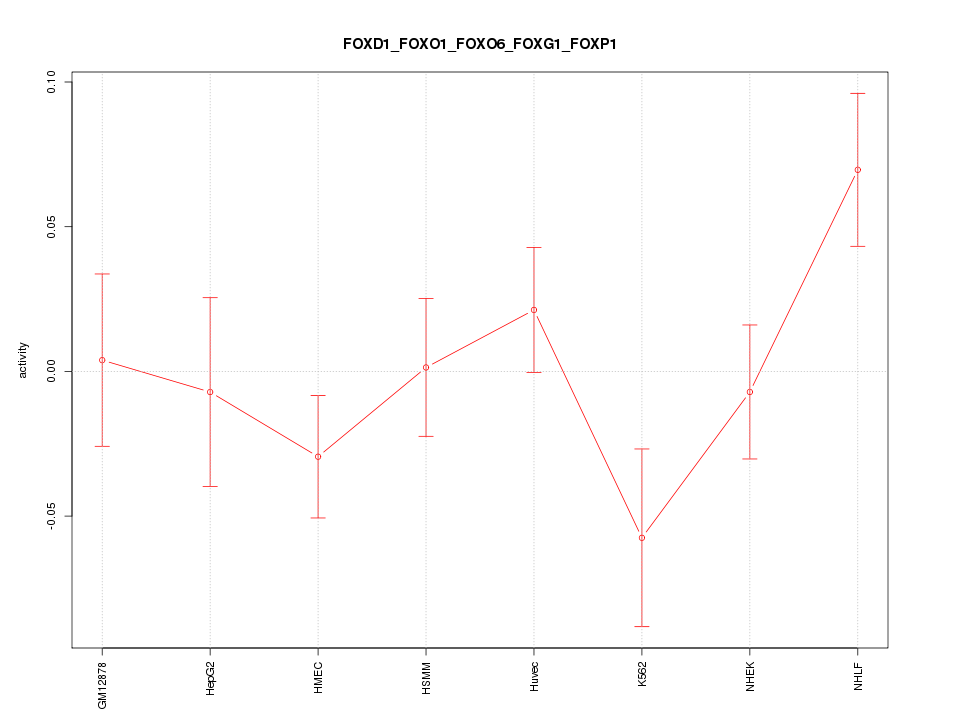
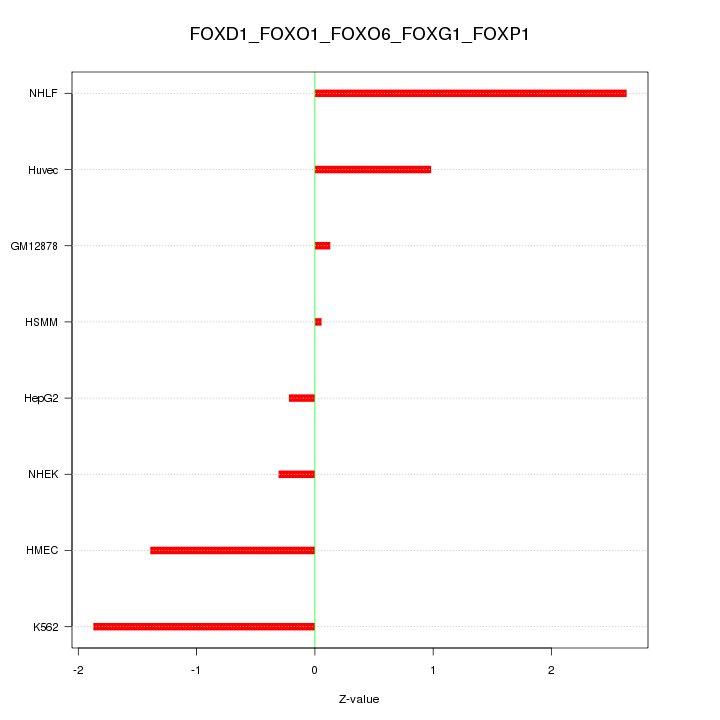
Motif ID: FOXD1_FOXO1_FOXO6_FOXG1_FOXP1
Z-value: 1.300
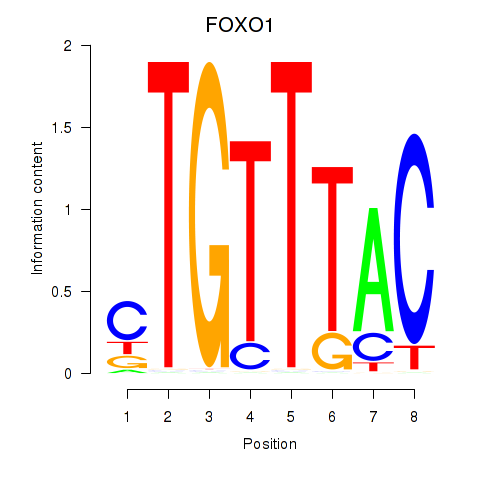
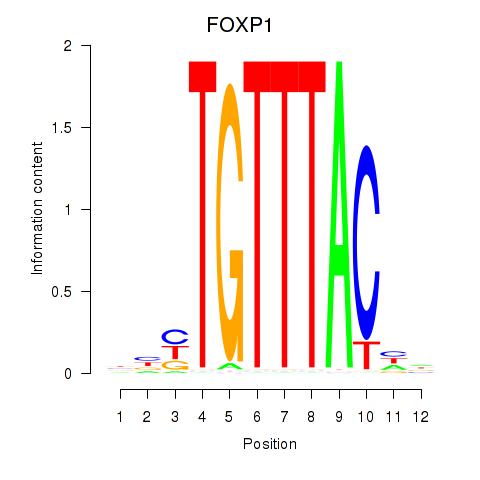
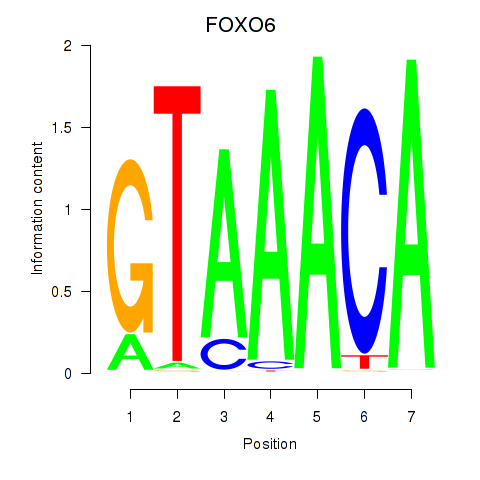
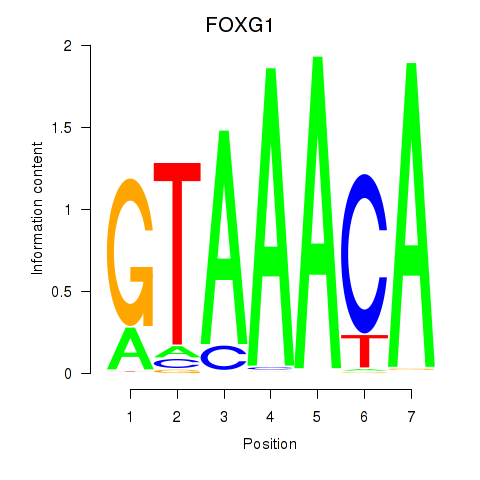
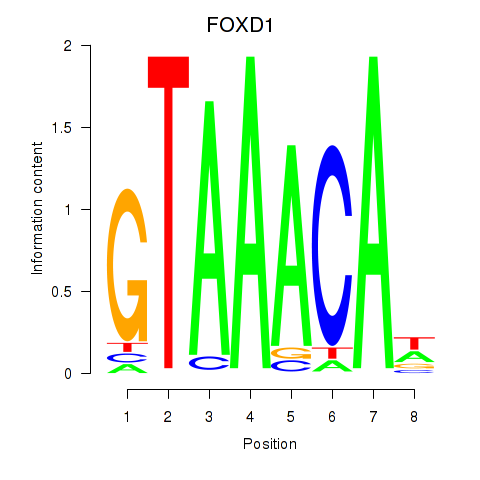
Transcription factors associated with FOXD1_FOXO1_FOXO6_FOXG1_FOXP1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| FOXD1 | ENSG00000251493.2 | FOXD1 |
| FOXG1 | ENSG00000176165.7 | FOXG1 |
| FOXO1 | ENSG00000150907.6 | FOXO1 |
| FOXO6 | ENSG00000204060.4 | FOXO6 |
| FOXP1 | ENSG00000114861.14 | FOXP1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.6 | 1.7 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.6 | 4.5 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.5 | 2.4 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.4 | 2.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.4 | 1.7 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.4 | 1.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.3 | 1.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.3 | 0.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 1.2 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.3 | 0.9 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.2 | 2.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 1.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 0.9 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.2 | 1.5 | GO:0060414 | aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 1.3 | GO:2000124 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 2.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.2 | 0.8 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.2 | 0.7 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.2 | 0.5 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.2 | 1.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.1 | 1.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.4 | GO:0060267 | transformation of host cell by virus(GO:0019087) positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.8 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 | 1.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 0.3 | GO:0014889 | muscle atrophy(GO:0014889) regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.1 | 0.9 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 0.4 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.1 | 1.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.4 | GO:0034587 | RNA 5'-end processing(GO:0000966) piRNA metabolic process(GO:0034587) |
| 0.1 | 0.6 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.3 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.1 | 2.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.2 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.6 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 2.0 | GO:0006911 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.1 | 0.4 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 0.3 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 | 0.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 0.2 | GO:0070544 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 0.5 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.1 | 1.7 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.1 | 0.7 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 | 0.2 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 1.2 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.2 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.0 | 0.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 1.3 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 5.2 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.4 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 15.1 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 1.1 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.9 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 0.9 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.4 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.2 | GO:0002051 | mesodermal cell fate commitment(GO:0001710) osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.2 | GO:0070245 | positive regulation of T cell apoptotic process(GO:0070234) positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 1.4 | GO:1900181 | negative regulation of protein import into nucleus(GO:0042308) negative regulation of protein localization to nucleus(GO:1900181) negative regulation of protein import(GO:1904590) |
| 0.0 | 2.1 | GO:0031668 | cellular response to extracellular stimulus(GO:0031668) |
| 0.0 | 0.5 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.2 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.6 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.8 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 0.1 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.2 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.3 | GO:0001502 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0060457 | negative regulation of digestive system process(GO:0060457) |
| 0.0 | 0.0 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.0 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.5 | GO:0030866 | cortical cytoskeleton organization(GO:0030865) cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.2 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 0.9 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 1.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.1 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.3 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.4 | GO:0070555 | response to interleukin-1(GO:0070555) |
| 0.0 | 0.0 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 5.1 | GO:0016049 | cell growth(GO:0016049) |
| 0.0 | 0.5 | GO:0007507 | heart development(GO:0007507) |
| 0.0 | 0.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.3 | GO:0050931 | melanocyte differentiation(GO:0030318) pigment cell differentiation(GO:0050931) |
| 0.0 | 0.0 | GO:0003071 | renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) |
| 0.0 | 0.6 | GO:0017156 | calcium ion regulated exocytosis(GO:0017156) |
| 0.0 | 0.6 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 2.8 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.2 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.2 | GO:0045446 | endothelial cell differentiation(GO:0045446) |
| 0.0 | 0.4 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.0 | 0.0 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.3 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.1 | GO:0000768 | syncytium formation by plasma membrane fusion(GO:0000768) syncytium formation(GO:0006949) myoblast fusion(GO:0007520) |
| 0.0 | 1.1 | GO:0036293 | response to hypoxia(GO:0001666) response to decreased oxygen levels(GO:0036293) |
| 0.0 | 1.7 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.6 | 1.9 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.3 | 1.7 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.3 | 0.9 | GO:0044422 | organelle part(GO:0044422) |
| 0.3 | 1.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.2 | 2.5 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 1.1 | GO:1990752 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.2 | 0.5 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) PAS complex(GO:0070772) |
| 0.1 | 0.7 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.4 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 1.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.4 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.1 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 2.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 0.4 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 1.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 11.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 4.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.6 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.4 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 3.3 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.0 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.2 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.7 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 2.5 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.6 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 1.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 5.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 4.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.7 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.0 | GO:0030141 | secretory granule(GO:0030141) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.5 | 2.4 | GO:0005534 | galactose binding(GO:0005534) |
| 0.5 | 6.0 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) RNA polymerase II basal transcription factor binding(GO:0001091) TFIIB-class transcription factor binding(GO:0001093) |
| 0.4 | 2.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) |
| 0.4 | 1.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.4 | 1.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 2.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.3 | 1.7 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.3 | 1.3 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.3 | 1.5 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 8.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 0.8 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 0.7 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.2 | 0.6 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 1.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 1.5 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.2 | 4.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 0.5 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.2 | 0.5 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.2 | 0.5 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.2 | 1.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 1.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 2.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.4 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.4 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.1 | 0.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 1.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.9 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 1.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.9 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.4 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 1.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.3 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.1 | 2.1 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.9 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.9 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 1.0 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.1 | 3.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.8 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.2 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.7 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 1.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0050833 | pyruvate secondary active transmembrane transporter activity(GO:0005477) pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 4.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.4 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0090079 | translation activator activity(GO:0008494) translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.0 | 0.4 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 4.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 2.1 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.4 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.4 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 3.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.4 | GO:0008186 | RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 1.6 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 1.8 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.0 | GO:0045030 | UTP-activated nucleotide receptor activity(GO:0045030) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.0 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.0 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.0 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 1.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.0 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.5 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.2 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.1 | ST_TYPE_I_INTERFERON_PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 0.5 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |