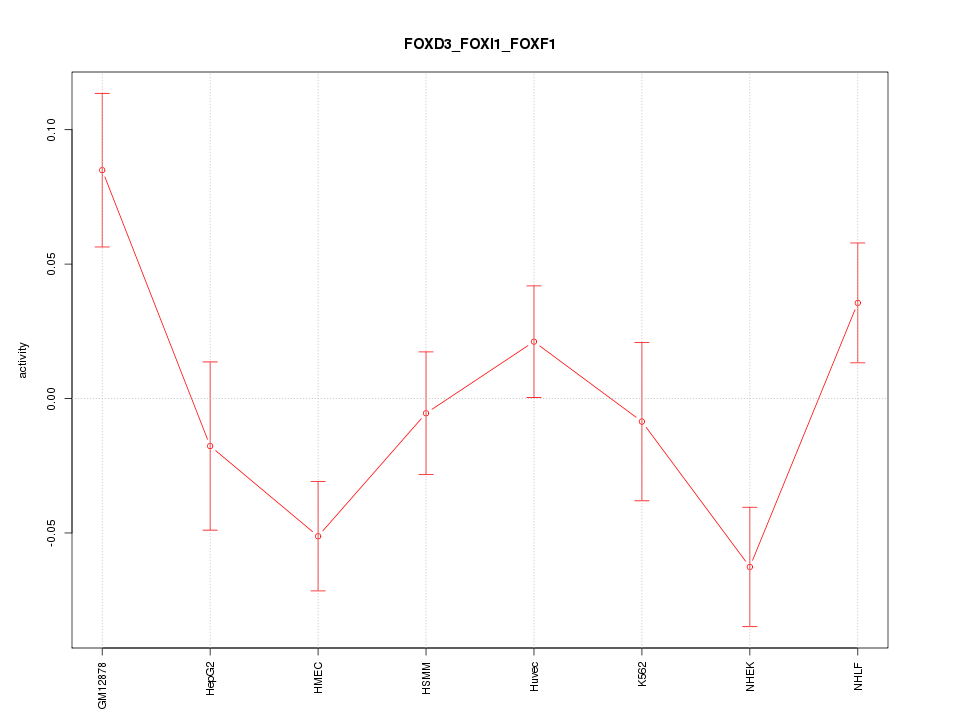
| 1.3 |
3.8 |
GO:0000738 |
DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.6 |
2.6 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 0.6 |
2.3 |
GO:0006848 |
pyruvate transport(GO:0006848) |
| 0.4 |
2.2 |
GO:0048014 |
Tie signaling pathway(GO:0048014) |
| 0.4 |
1.3 |
GO:0071380 |
cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.4 |
2.6 |
GO:0045627 |
regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.3 |
1.0 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.3 |
1.9 |
GO:0006772 |
thiamine metabolic process(GO:0006772) |
| 0.3 |
0.6 |
GO:0006154 |
adenosine catabolic process(GO:0006154) |
| 0.3 |
1.8 |
GO:0043316 |
natural killer cell activation involved in immune response(GO:0002323) cytotoxic T cell degranulation(GO:0043316) natural killer cell degranulation(GO:0043320) |
| 0.3 |
1.7 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.3 |
0.8 |
GO:0070340 |
detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.3 |
1.6 |
GO:2000124 |
cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 |
1.6 |
GO:0006658 |
phosphatidylserine metabolic process(GO:0006658) |
| 0.2 |
0.6 |
GO:0060318 |
definitive erythrocyte differentiation(GO:0060318) |
| 0.2 |
1.8 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.2 |
1.6 |
GO:0060100 |
regulation of phagocytosis, engulfment(GO:0060099) positive regulation of phagocytosis, engulfment(GO:0060100) regulation of membrane invagination(GO:1905153) positive regulation of membrane invagination(GO:1905155) |
| 0.2 |
0.6 |
GO:2000121 |
regulation of removal of superoxide radicals(GO:2000121) |
| 0.2 |
1.6 |
GO:0050857 |
positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.2 |
0.9 |
GO:0018106 |
peptidyl-histidine phosphorylation(GO:0018106) |
| 0.2 |
0.7 |
GO:0002313 |
mature B cell differentiation involved in immune response(GO:0002313) |
| 0.2 |
0.5 |
GO:0046586 |
regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 |
1.4 |
GO:0090292 |
nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 |
0.7 |
GO:0043306 |
positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 |
0.4 |
GO:0033578 |
protein glycosylation in Golgi(GO:0033578) |
| 0.1 |
0.4 |
GO:0045601 |
regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 |
0.4 |
GO:0001757 |
somite specification(GO:0001757) |
| 0.1 |
0.5 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 |
0.9 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.1 |
0.7 |
GO:0043402 |
glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 |
0.5 |
GO:0046121 |
deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 |
1.4 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
0.4 |
GO:0014831 |
intestine smooth muscle contraction(GO:0014827) gastro-intestinal system smooth muscle contraction(GO:0014831) |
| 0.1 |
0.5 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 |
1.4 |
GO:0006975 |
DNA damage induced protein phosphorylation(GO:0006975) |
| 0.1 |
0.5 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 |
0.1 |
GO:0045744 |
negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 |
0.6 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.1 |
0.5 |
GO:0071898 |
regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.1 |
0.3 |
GO:0071460 |
cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 |
0.3 |
GO:0006288 |
base-excision repair, DNA ligation(GO:0006288) |
| 0.1 |
0.5 |
GO:0032497 |
detection of lipopolysaccharide(GO:0032497) |
| 0.1 |
1.5 |
GO:0015939 |
pantothenate metabolic process(GO:0015939) |
| 0.1 |
0.6 |
GO:0019722 |
calcium-mediated signaling(GO:0019722) |
| 0.1 |
0.3 |
GO:0046292 |
formaldehyde metabolic process(GO:0046292) |
| 0.1 |
0.8 |
GO:0040037 |
negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.1 |
0.5 |
GO:0046642 |
negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 |
0.7 |
GO:0043931 |
ossification involved in bone maturation(GO:0043931) |
| 0.1 |
0.6 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 |
0.4 |
GO:0045759 |
negative regulation of action potential(GO:0045759) |
| 0.1 |
0.3 |
GO:1903961 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.1 |
0.2 |
GO:0006816 |
calcium ion transport(GO:0006816) |
| 0.1 |
0.3 |
GO:0030101 |
natural killer cell activation(GO:0030101) |
| 0.1 |
0.1 |
GO:0010248 |
establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.1 |
0.8 |
GO:0008354 |
germ cell migration(GO:0008354) |
| 0.1 |
0.4 |
GO:0051571 |
positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.1 |
0.4 |
GO:0010771 |
negative regulation of cell morphogenesis involved in differentiation(GO:0010771) |
| 0.1 |
0.2 |
GO:0090598 |
male genitalia morphogenesis(GO:0048808) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) male anatomical structure morphogenesis(GO:0090598) |
| 0.1 |
0.3 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 |
1.4 |
GO:0006911 |
phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.1 |
0.2 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.1 |
0.2 |
GO:0042320 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) |
| 0.1 |
0.2 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.1 |
8.5 |
GO:0006367 |
transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 |
0.9 |
GO:0019228 |
neuronal action potential(GO:0019228) |
| 0.1 |
0.2 |
GO:0070171 |
negative regulation of tooth mineralization(GO:0070171) |
| 0.1 |
0.5 |
GO:0035162 |
embryonic hemopoiesis(GO:0035162) |
| 0.1 |
0.2 |
GO:0007285 |
primary spermatocyte growth(GO:0007285) |
| 0.1 |
0.1 |
GO:0060398 |
regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.1 |
0.3 |
GO:0071168 |
protein localization to chromatin(GO:0071168) |
| 0.1 |
0.7 |
GO:0006002 |
fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 |
1.1 |
GO:0001502 |
cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.1 |
0.1 |
GO:0050766 |
regulation of phagocytosis(GO:0050764) positive regulation of phagocytosis(GO:0050766) |
| 0.1 |
0.3 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.1 |
0.4 |
GO:0045059 |
positive thymic T cell selection(GO:0045059) |
| 0.1 |
1.6 |
GO:0002286 |
T cell activation involved in immune response(GO:0002286) |
| 0.1 |
0.1 |
GO:0002523 |
leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 |
0.1 |
GO:0072678 |
T cell chemotaxis(GO:0010818) regulation of T cell chemotaxis(GO:0010819) positive regulation of T cell chemotaxis(GO:0010820) T cell migration(GO:0072678) regulation of lymphocyte chemotaxis(GO:1901623) regulation of lymphocyte migration(GO:2000401) positive regulation of lymphocyte migration(GO:2000403) regulation of T cell migration(GO:2000404) positive regulation of T cell migration(GO:2000406) |
| 0.1 |
0.1 |
GO:0046639 |
negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.1 |
0.2 |
GO:0009106 |
lipoate metabolic process(GO:0009106) |
| 0.1 |
1.3 |
GO:0050830 |
defense response to Gram-positive bacterium(GO:0050830) |
| 0.1 |
0.2 |
GO:0043095 |
regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.1 |
0.4 |
GO:0001554 |
luteolysis(GO:0001554) |
| 0.1 |
0.6 |
GO:0000076 |
DNA replication checkpoint(GO:0000076) |
| 0.1 |
1.1 |
GO:0042531 |
positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 |
0.5 |
GO:0071692 |
sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 |
0.3 |
GO:0030951 |
establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 |
0.2 |
GO:0009162 |
deoxyribonucleoside monophosphate metabolic process(GO:0009162) pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) dUMP metabolic process(GO:0046078) |
| 0.0 |
0.3 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.0 |
0.2 |
GO:0019276 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 |
0.1 |
GO:0007207 |
phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 |
0.3 |
GO:0044245 |
polysaccharide digestion(GO:0044245) |
| 0.0 |
0.3 |
GO:0070245 |
positive regulation of T cell apoptotic process(GO:0070234) positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 |
0.2 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) |
| 0.0 |
0.2 |
GO:0046952 |
ketone body catabolic process(GO:0046952) |
| 0.0 |
0.1 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 |
0.2 |
GO:0090025 |
regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 |
2.7 |
GO:0097696 |
JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.0 |
1.2 |
GO:0001508 |
action potential(GO:0001508) |
| 0.0 |
0.3 |
GO:0007598 |
blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 |
0.5 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) |
| 0.0 |
0.2 |
GO:0045953 |
desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) negative regulation of natural killer cell mediated immunity(GO:0002716) negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.0 |
0.8 |
GO:0000303 |
response to superoxide(GO:0000303) |
| 0.0 |
0.4 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.0 |
0.8 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.6 |
GO:0045599 |
negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 |
0.8 |
GO:0000380 |
alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 |
0.2 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.0 |
1.1 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
0.1 |
GO:0052805 |
histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 |
0.2 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 |
0.3 |
GO:0031274 |
pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 |
0.1 |
GO:0030202 |
heparin metabolic process(GO:0030202) |
| 0.0 |
2.1 |
GO:0030183 |
B cell differentiation(GO:0030183) |
| 0.0 |
0.3 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
0.1 |
GO:0001692 |
histamine metabolic process(GO:0001692) |
| 0.0 |
1.1 |
GO:0030866 |
cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 |
0.2 |
GO:0009128 |
AMP catabolic process(GO:0006196) nucleoside monophosphate catabolic process(GO:0009125) purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 |
0.2 |
GO:0045916 |
negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 |
0.1 |
GO:0001831 |
trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 |
0.2 |
GO:0042635 |
positive regulation of hair cycle(GO:0042635) regulation of hair follicle development(GO:0051797) positive regulation of hair follicle development(GO:0051798) |
| 0.0 |
0.1 |
GO:0044240 |
multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 |
2.4 |
GO:0061025 |
membrane fusion(GO:0061025) |
| 0.0 |
0.5 |
GO:0009154 |
cAMP catabolic process(GO:0006198) purine ribonucleotide catabolic process(GO:0009154) |
| 0.0 |
0.6 |
GO:0060334 |
regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 |
0.3 |
GO:0031000 |
response to caffeine(GO:0031000) |
| 0.0 |
0.1 |
GO:0050968 |
detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 |
0.1 |
GO:0009301 |
snRNA transcription(GO:0009301) |
| 0.0 |
0.7 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.0 |
0.6 |
GO:1902749 |
regulation of G2/M transition of mitotic cell cycle(GO:0010389) regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 |
0.1 |
GO:0008295 |
spermidine biosynthetic process(GO:0008295) |
| 0.0 |
0.1 |
GO:0007182 |
common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 |
0.2 |
GO:1901503 |
ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 |
0.9 |
GO:0050931 |
melanocyte differentiation(GO:0030318) pigment cell differentiation(GO:0050931) |
| 0.0 |
0.3 |
GO:0050856 |
regulation of T cell receptor signaling pathway(GO:0050856) |
| 0.0 |
0.4 |
GO:0070206 |
protein trimerization(GO:0070206) |
| 0.0 |
0.1 |
GO:0071462 |
cellular response to water stimulus(GO:0071462) |
| 0.0 |
0.2 |
GO:0007346 |
regulation of mitotic cell cycle(GO:0007346) |
| 0.0 |
0.0 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.0 |
0.2 |
GO:0031122 |
cytoplasmic microtubule organization(GO:0031122) |
| 0.0 |
0.8 |
GO:0051693 |
actin filament capping(GO:0051693) |
| 0.0 |
0.1 |
GO:0071635 |
negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 |
0.1 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.0 |
0.0 |
GO:0032693 |
tolerance induction to self antigen(GO:0002513) regulation of chronic inflammatory response(GO:0002676) negative regulation of chronic inflammatory response(GO:0002677) negative regulation of interleukin-10 production(GO:0032693) positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 |
0.2 |
GO:0048853 |
forebrain morphogenesis(GO:0048853) |
| 0.0 |
0.1 |
GO:0000710 |
meiotic mismatch repair(GO:0000710) |
| 0.0 |
0.3 |
GO:0045954 |
positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 |
0.1 |
GO:0060291 |
long-term synaptic potentiation(GO:0060291) |
| 0.0 |
0.1 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
0.4 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 |
0.1 |
GO:0046784 |
viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 |
0.0 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.0 |
0.3 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 |
0.1 |
GO:0006386 |
transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 |
1.0 |
GO:0007193 |
adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 |
0.2 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.0 |
0.1 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 |
0.1 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 |
0.3 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.1 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.3 |
GO:0048661 |
positive regulation of smooth muscle cell proliferation(GO:0048661) |
| 0.0 |
0.1 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 |
0.0 |
GO:0010430 |
ethanol catabolic process(GO:0006068) fatty acid omega-oxidation(GO:0010430) |
| 0.0 |
0.1 |
GO:0032414 |
positive regulation of ion transmembrane transporter activity(GO:0032414) |
| 0.0 |
0.1 |
GO:0006422 |
aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 |
0.1 |
GO:0031643 |
positive regulation of myelination(GO:0031643) |
| 0.0 |
0.2 |
GO:0071496 |
cellular response to external stimulus(GO:0071496) |
| 0.0 |
0.9 |
GO:0048704 |
embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 |
0.3 |
GO:0021846 |
cell proliferation in forebrain(GO:0021846) |
| 0.0 |
0.2 |
GO:0051924 |
regulation of calcium ion transport(GO:0051924) |
| 0.0 |
0.1 |
GO:0015793 |
glycerol transport(GO:0015793) |
| 0.0 |
0.1 |
GO:0046470 |
phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 |
1.3 |
GO:0006977 |
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) positive regulation of cell cycle arrest(GO:0071158) signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.0 |
0.1 |
GO:1902170 |
cellular response to nitric oxide(GO:0071732) cellular response to reactive nitrogen species(GO:1902170) |
| 0.0 |
0.1 |
GO:0010815 |
bradykinin catabolic process(GO:0010815) beta-amyloid metabolic process(GO:0050435) |
| 0.0 |
0.1 |
GO:0042074 |
cell migration involved in gastrulation(GO:0042074) |
| 0.0 |
0.5 |
GO:0008284 |
positive regulation of cell proliferation(GO:0008284) |
| 0.0 |
0.0 |
GO:0033119 |
negative regulation of RNA splicing(GO:0033119) |
| 0.0 |
0.3 |
GO:0000080 |
mitotic G1 phase(GO:0000080) |
| 0.0 |
0.2 |
GO:0007194 |
negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) |
| 0.0 |
0.5 |
GO:0032480 |
negative regulation of type I interferon production(GO:0032480) |
| 0.0 |
0.1 |
GO:0007219 |
Notch signaling pathway(GO:0007219) |
| 0.0 |
0.8 |
GO:0006338 |
chromatin remodeling(GO:0006338) |
| 0.0 |
0.6 |
GO:0017156 |
calcium ion regulated exocytosis(GO:0017156) |
| 0.0 |
0.8 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.0 |
0.2 |
GO:0070374 |
positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.0 |
0.0 |
GO:0031641 |
regulation of myelination(GO:0031641) |
| 0.0 |
0.2 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.0 |
0.0 |
GO:0048024 |
regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 |
0.1 |
GO:0007080 |
mitotic metaphase plate congression(GO:0007080) |
| 0.0 |
0.4 |
GO:0000725 |
double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 |
0.0 |
GO:0072367 |
regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 |
0.1 |
GO:0042226 |
interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 |
2.0 |
GO:0050900 |
leukocyte migration(GO:0050900) |
| 0.0 |
0.0 |
GO:1901142 |
insulin processing(GO:0030070) insulin metabolic process(GO:1901142) |