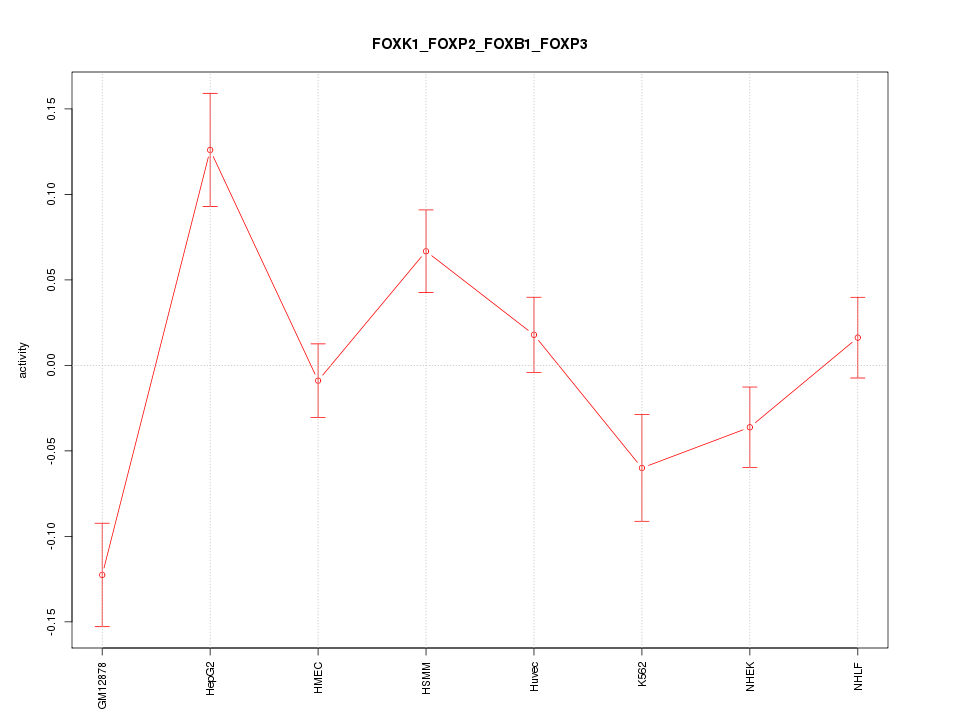
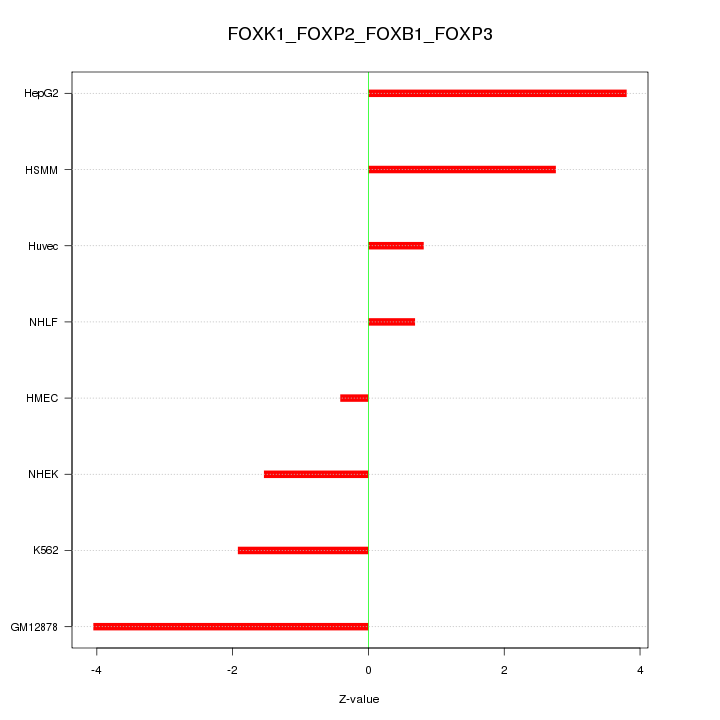
| 4.0 |
11.9 |
GO:0016554 |
cytidine to uridine editing(GO:0016554) |
| 2.4 |
9.4 |
GO:0048619 |
embryonic hindgut morphogenesis(GO:0048619) |
| 2.2 |
6.7 |
GO:2000910 |
regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 1.6 |
4.8 |
GO:0046010 |
positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 1.3 |
10.5 |
GO:0007598 |
blood coagulation, extrinsic pathway(GO:0007598) |
| 1.2 |
1.2 |
GO:0010903 |
negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 1.2 |
7.0 |
GO:0045916 |
negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 1.1 |
10.0 |
GO:0031639 |
plasminogen activation(GO:0031639) |
| 1.1 |
4.3 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.8 |
4.0 |
GO:0006651 |
diacylglycerol biosynthetic process(GO:0006651) |
| 0.8 |
2.3 |
GO:0052805 |
histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.7 |
7.7 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.7 |
2.8 |
GO:0010716 |
regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.7 |
0.7 |
GO:0050918 |
positive chemotaxis(GO:0050918) |
| 0.7 |
2.0 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.6 |
1.9 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.6 |
1.8 |
GO:0071380 |
cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.6 |
1.8 |
GO:0014889 |
muscle atrophy(GO:0014889) regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.6 |
2.3 |
GO:0060298 |
positive regulation of sarcomere organization(GO:0060298) |
| 0.5 |
1.6 |
GO:0006015 |
5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.5 |
3.1 |
GO:0060242 |
contact inhibition(GO:0060242) |
| 0.5 |
1.4 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.5 |
1.8 |
GO:0010727 |
negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.5 |
2.3 |
GO:0050703 |
interleukin-1 alpha secretion(GO:0050703) |
| 0.4 |
3.9 |
GO:0040037 |
negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.4 |
1.7 |
GO:0009609 |
response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.4 |
1.3 |
GO:1901142 |
insulin processing(GO:0030070) insulin metabolic process(GO:1901142) |
| 0.4 |
1.9 |
GO:0060414 |
aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.4 |
2.2 |
GO:0007525 |
somatic muscle development(GO:0007525) |
| 0.4 |
1.4 |
GO:0060457 |
negative regulation of digestive system process(GO:0060457) |
| 0.4 |
1.4 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.3 |
0.3 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.3 |
18.7 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.3 |
4.7 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.3 |
1.3 |
GO:0071428 |
rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.3 |
4.4 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.3 |
2.9 |
GO:0010766 |
negative regulation of sodium ion transport(GO:0010766) |
| 0.3 |
1.3 |
GO:0072180 |
mesonephric duct morphogenesis(GO:0072180) |
| 0.3 |
0.9 |
GO:0003350 |
pulmonary myocardium development(GO:0003350) |
| 0.3 |
0.9 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.3 |
1.2 |
GO:0001705 |
ectoderm formation(GO:0001705) central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.3 |
1.4 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.3 |
2.8 |
GO:0031641 |
regulation of myelination(GO:0031641) |
| 0.3 |
0.8 |
GO:0070544 |
regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) histone H3-K36 demethylation(GO:0070544) |
| 0.3 |
2.7 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.3 |
1.6 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.3 |
0.8 |
GO:0019521 |
aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.2 |
3.7 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.2 |
1.5 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.2 |
0.7 |
GO:0001544 |
initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.2 |
2.6 |
GO:0006907 |
pinocytosis(GO:0006907) |
| 0.2 |
0.9 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.2 |
2.6 |
GO:0042447 |
hormone catabolic process(GO:0042447) |
| 0.2 |
0.2 |
GO:0019184 |
nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.2 |
2.1 |
GO:0060762 |
regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.2 |
0.6 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 0.2 |
1.8 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 |
0.6 |
GO:0090086 |
negative regulation of protein deubiquitination(GO:0090086) |
| 0.2 |
5.7 |
GO:1900047 |
negative regulation of blood coagulation(GO:0030195) negative regulation of wound healing(GO:0061045) negative regulation of hemostasis(GO:1900047) |
| 0.2 |
1.5 |
GO:0008354 |
germ cell migration(GO:0008354) |
| 0.2 |
1.6 |
GO:0097435 |
extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.2 |
1.2 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.2 |
1.0 |
GO:0070245 |
positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.2 |
0.7 |
GO:0045872 |
positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.2 |
0.3 |
GO:0007499 |
ectoderm and mesoderm interaction(GO:0007499) |
| 0.2 |
1.7 |
GO:0032000 |
positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 |
0.3 |
GO:0045750 |
obsolete positive regulation of S phase of mitotic cell cycle(GO:0045750) |
| 0.2 |
0.5 |
GO:0019731 |
antibacterial humoral response(GO:0019731) |
| 0.2 |
2.2 |
GO:0008211 |
glucocorticoid metabolic process(GO:0008211) |
| 0.2 |
1.5 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.2 |
1.1 |
GO:0070779 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 |
2.1 |
GO:1904893 |
negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 |
1.4 |
GO:0018126 |
protein hydroxylation(GO:0018126) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.2 |
1.4 |
GO:0032528 |
microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.2 |
0.5 |
GO:0007231 |
osmosensory signaling pathway(GO:0007231) |
| 0.1 |
0.6 |
GO:0071400 |
cellular response to oleic acid(GO:0071400) |
| 0.1 |
0.6 |
GO:0007368 |
determination of left/right symmetry(GO:0007368) specification of symmetry(GO:0009799) determination of bilateral symmetry(GO:0009855) |
| 0.1 |
0.6 |
GO:0015825 |
L-serine transport(GO:0015825) |
| 0.1 |
0.4 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 |
1.7 |
GO:0046323 |
glucose import(GO:0046323) |
| 0.1 |
3.1 |
GO:0046627 |
negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 |
0.6 |
GO:0060209 |
estrus(GO:0060209) |
| 0.1 |
6.0 |
GO:0008206 |
bile acid metabolic process(GO:0008206) |
| 0.1 |
0.1 |
GO:0046642 |
negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 |
0.4 |
GO:0060574 |
trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) intestinal epithelial cell maturation(GO:0060574) intestinal epithelial cell development(GO:0060576) |
| 0.1 |
9.3 |
GO:0055088 |
lipid homeostasis(GO:0055088) |
| 0.1 |
0.5 |
GO:0000966 |
RNA 5'-end processing(GO:0000966) |
| 0.1 |
0.9 |
GO:0033089 |
positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 |
1.1 |
GO:0042448 |
progesterone metabolic process(GO:0042448) |
| 0.1 |
1.5 |
GO:0051597 |
response to methylmercury(GO:0051597) |
| 0.1 |
0.5 |
GO:0034331 |
cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.1 |
0.7 |
GO:0042693 |
muscle cell fate commitment(GO:0042693) |
| 0.1 |
0.4 |
GO:0042416 |
dopamine biosynthetic process(GO:0042416) |
| 0.1 |
1.1 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 |
1.9 |
GO:0008210 |
estrogen metabolic process(GO:0008210) |
| 0.1 |
0.9 |
GO:0003065 |
regulation of heart rate by chemical signal(GO:0003062) positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 |
0.8 |
GO:0045071 |
negative regulation of viral genome replication(GO:0045071) |
| 0.1 |
0.3 |
GO:0032074 |
negative regulation of nuclease activity(GO:0032074) |
| 0.1 |
1.1 |
GO:0046473 |
phosphatidic acid metabolic process(GO:0046473) |
| 0.1 |
1.9 |
GO:0031000 |
response to caffeine(GO:0031000) |
| 0.1 |
0.6 |
GO:0061003 |
positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 |
3.0 |
GO:0034199 |
activation of protein kinase A activity(GO:0034199) |
| 0.1 |
1.5 |
GO:0006787 |
porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 |
37.1 |
GO:0006936 |
muscle contraction(GO:0006936) |
| 0.1 |
1.1 |
GO:0045070 |
positive regulation of viral genome replication(GO:0045070) |
| 0.1 |
1.0 |
GO:0050872 |
white fat cell differentiation(GO:0050872) |
| 0.1 |
0.4 |
GO:0046621 |
negative regulation of organ growth(GO:0046621) |
| 0.1 |
0.4 |
GO:0060839 |
lymphatic endothelial cell fate commitment(GO:0060838) endothelial cell fate commitment(GO:0060839) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 |
0.9 |
GO:0042594 |
response to starvation(GO:0042594) |
| 0.1 |
0.8 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.1 |
0.3 |
GO:0051088 |
PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 |
1.8 |
GO:0030199 |
collagen fibril organization(GO:0030199) |
| 0.1 |
0.7 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.1 |
0.2 |
GO:0045214 |
sarcomere organization(GO:0045214) |
| 0.1 |
2.0 |
GO:0008347 |
glial cell migration(GO:0008347) |
| 0.1 |
0.8 |
GO:0007567 |
parturition(GO:0007567) |
| 0.1 |
0.7 |
GO:0042475 |
odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 |
2.5 |
GO:0015914 |
phospholipid transport(GO:0015914) |
| 0.1 |
0.7 |
GO:0050847 |
progesterone receptor signaling pathway(GO:0050847) |
| 0.1 |
0.3 |
GO:0034435 |
regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.1 |
0.2 |
GO:0042905 |
9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 |
5.5 |
GO:0051592 |
response to calcium ion(GO:0051592) |
| 0.1 |
2.5 |
GO:0030325 |
adrenal gland development(GO:0030325) |
| 0.1 |
1.7 |
GO:0006825 |
copper ion transport(GO:0006825) |
| 0.1 |
0.3 |
GO:0009822 |
alkaloid catabolic process(GO:0009822) |
| 0.1 |
0.2 |
GO:0045723 |
positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 |
0.6 |
GO:0006553 |
lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.1 |
1.6 |
GO:0045104 |
intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 |
0.3 |
GO:0001823 |
ureteric bud development(GO:0001657) mesonephros development(GO:0001823) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) |
| 0.1 |
0.5 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.1 |
0.3 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.1 |
0.2 |
GO:0009756 |
carbohydrate mediated signaling(GO:0009756) |
| 0.1 |
0.6 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.1 |
0.1 |
GO:0019087 |
transformation of host cell by virus(GO:0019087) |
| 0.1 |
0.3 |
GO:0021520 |
spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 |
0.3 |
GO:0072383 |
plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 |
0.6 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.1 |
0.3 |
GO:1903301 |
regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 |
0.2 |
GO:0050758 |
thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 |
0.2 |
GO:0009820 |
alkaloid metabolic process(GO:0009820) |
| 0.1 |
0.1 |
GO:1901522 |
positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.1 |
1.0 |
GO:0045671 |
negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 |
0.2 |
GO:0043162 |
ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 |
0.3 |
GO:0015871 |
choline transport(GO:0015871) |
| 0.1 |
0.6 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 |
4.2 |
GO:0031668 |
cellular response to extracellular stimulus(GO:0031668) |
| 0.1 |
5.8 |
GO:0051384 |
response to glucocorticoid(GO:0051384) |
| 0.1 |
0.3 |
GO:0046121 |
deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 |
0.1 |
GO:0033629 |
negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.1 |
1.4 |
GO:0006693 |
prostaglandin metabolic process(GO:0006693) |
| 0.1 |
0.9 |
GO:0042403 |
thyroid hormone metabolic process(GO:0042403) |
| 0.1 |
0.3 |
GO:0032933 |
response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 |
0.3 |
GO:0032570 |
response to progesterone(GO:0032570) |
| 0.1 |
0.2 |
GO:0043922 |
negative regulation by host of viral transcription(GO:0043922) |
| 0.1 |
0.1 |
GO:0002220 |
innate immune response activating cell surface receptor signaling pathway(GO:0002220) |
| 0.1 |
0.2 |
GO:0014067 |
negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 |
2.9 |
GO:0009749 |
response to glucose(GO:0009749) |
| 0.1 |
0.2 |
GO:0070584 |
mitochondrion morphogenesis(GO:0070584) |
| 0.1 |
0.2 |
GO:0014912 |
negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.1 |
0.1 |
GO:0070309 |
lens fiber cell morphogenesis(GO:0070309) |
| 0.1 |
0.2 |
GO:0002667 |
lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 |
0.2 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.1 |
0.3 |
GO:0051281 |
positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.1 |
0.7 |
GO:0071445 |
obsolete cellular response to protein stimulus(GO:0071445) |
| 0.1 |
0.1 |
GO:0002523 |
leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 |
0.4 |
GO:0010799 |
regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 |
0.3 |
GO:0032878 |
regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 |
0.5 |
GO:0015780 |
nucleotide-sugar transport(GO:0015780) |
| 0.1 |
4.2 |
GO:0008360 |
regulation of cell shape(GO:0008360) |
| 0.1 |
0.2 |
GO:0042519 |
tyrosine phosphorylation of Stat4 protein(GO:0042504) regulation of tyrosine phosphorylation of Stat4 protein(GO:0042519) positive regulation of tyrosine phosphorylation of Stat4 protein(GO:0042520) |
| 0.1 |
0.6 |
GO:0050687 |
negative regulation of defense response to virus(GO:0050687) |
| 0.0 |
0.3 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
1.0 |
GO:0007045 |
cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 |
1.8 |
GO:0033572 |
ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 |
0.3 |
GO:0046618 |
drug export(GO:0046618) |
| 0.0 |
8.7 |
GO:0006979 |
response to oxidative stress(GO:0006979) |
| 0.0 |
0.1 |
GO:0051083 |
'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 |
1.1 |
GO:0009083 |
branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
1.6 |
GO:0017148 |
negative regulation of translation(GO:0017148) |
| 0.0 |
0.3 |
GO:0002021 |
response to dietary excess(GO:0002021) |
| 0.0 |
1.0 |
GO:0042994 |
cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 |
0.0 |
GO:1900087 |
traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle phase transition(GO:1901989) positive regulation of mitotic cell cycle phase transition(GO:1901992) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.0 |
0.2 |
GO:0002032 |
desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 |
0.1 |
GO:0050747 |
positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 |
1.4 |
GO:0042990 |
regulation of transcription factor import into nucleus(GO:0042990) transcription factor import into nucleus(GO:0042991) |
| 0.0 |
0.2 |
GO:0001575 |
globoside metabolic process(GO:0001575) |
| 0.0 |
0.2 |
GO:0000271 |
polysaccharide biosynthetic process(GO:0000271) glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) cellular polysaccharide biosynthetic process(GO:0033692) |
| 0.0 |
0.7 |
GO:0098764 |
meiosis I cell cycle phase(GO:0098764) |
| 0.0 |
1.6 |
GO:0030837 |
negative regulation of actin filament polymerization(GO:0030837) |
| 0.0 |
0.1 |
GO:0010896 |
regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 |
0.2 |
GO:0042770 |
DNA damage response, signal transduction by p53 class mediator(GO:0030330) signal transduction in response to DNA damage(GO:0042770) |
| 0.0 |
0.2 |
GO:0050884 |
neuromuscular process controlling posture(GO:0050884) |
| 0.0 |
1.6 |
GO:0006749 |
glutathione metabolic process(GO:0006749) |
| 0.0 |
0.3 |
GO:0044341 |
sodium-dependent phosphate transport(GO:0044341) |
| 0.0 |
0.1 |
GO:0010966 |
regulation of phosphate transport(GO:0010966) |
| 0.0 |
0.2 |
GO:0007584 |
response to nutrient(GO:0007584) |
| 0.0 |
0.0 |
GO:0002921 |
negative regulation of humoral immune response(GO:0002921) |
| 0.0 |
0.2 |
GO:0032754 |
positive regulation of interleukin-5 production(GO:0032754) |
| 0.0 |
1.1 |
GO:0001525 |
angiogenesis(GO:0001525) |
| 0.0 |
0.2 |
GO:0097061 |
dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 |
0.6 |
GO:0006518 |
peptide metabolic process(GO:0006518) |
| 0.0 |
1.3 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 |
1.1 |
GO:0007043 |
cell-cell junction assembly(GO:0007043) |
| 0.0 |
0.3 |
GO:0006910 |
phagocytosis, recognition(GO:0006910) |
| 0.0 |
0.1 |
GO:0007181 |
transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 |
0.9 |
GO:0015949 |
nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 |
0.1 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 |
0.5 |
GO:0016572 |
histone phosphorylation(GO:0016572) |
| 0.0 |
1.1 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.0 |
0.2 |
GO:0090292 |
nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 |
0.6 |
GO:0002576 |
platelet degranulation(GO:0002576) |
| 0.0 |
0.4 |
GO:0051881 |
regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 |
0.3 |
GO:0019885 |
antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 |
0.2 |
GO:0009190 |
cyclic nucleotide biosynthetic process(GO:0009190) cyclic purine nucleotide metabolic process(GO:0052652) |
| 0.0 |
0.5 |
GO:0030968 |
endoplasmic reticulum unfolded protein response(GO:0030968) cellular response to unfolded protein(GO:0034620) |
| 0.0 |
1.4 |
GO:0010038 |
response to metal ion(GO:0010038) |
| 0.0 |
0.1 |
GO:0006662 |
glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 |
0.1 |
GO:0043267 |
negative regulation of potassium ion transport(GO:0043267) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 |
0.1 |
GO:0032914 |
positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 |
0.4 |
GO:0030032 |
lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 |
0.1 |
GO:0032747 |
positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 |
0.1 |
GO:0021984 |
adenohypophysis development(GO:0021984) |
| 0.0 |
1.5 |
GO:0030198 |
extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 |
0.1 |
GO:0060164 |
regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 |
2.6 |
GO:0010951 |
negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 |
0.8 |
GO:0045727 |
positive regulation of cellular amide metabolic process(GO:0034250) positive regulation of translation(GO:0045727) |
| 0.0 |
0.0 |
GO:0097352 |
autophagosome maturation(GO:0097352) |
| 0.0 |
1.2 |
GO:0045165 |
cell fate commitment(GO:0045165) |
| 0.0 |
0.1 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.0 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.0 |
0.4 |
GO:0010633 |
negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 |
0.7 |
GO:0030866 |
cortical cytoskeleton organization(GO:0030865) cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 |
0.5 |
GO:0006691 |
leukotriene metabolic process(GO:0006691) |
| 0.0 |
0.1 |
GO:0008634 |
obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.0 |
0.4 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.0 |
0.2 |
GO:0045744 |
negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 |
0.0 |
GO:0070307 |
lens fiber cell development(GO:0070307) |
| 0.0 |
0.1 |
GO:0043923 |
positive regulation by host of viral transcription(GO:0043923) |
| 0.0 |
0.6 |
GO:0043044 |
ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 |
0.1 |
GO:0007204 |
positive regulation of cytosolic calcium ion concentration(GO:0007204) |
| 0.0 |
0.2 |
GO:0006998 |
nuclear envelope organization(GO:0006998) |
| 0.0 |
2.3 |
GO:0006814 |
sodium ion transport(GO:0006814) |
| 0.0 |
0.2 |
GO:0008272 |
sulfate transport(GO:0008272) |
| 0.0 |
0.2 |
GO:0045749 |
obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 |
1.0 |
GO:0051297 |
microtubule organizing center organization(GO:0031023) centrosome organization(GO:0051297) |
| 0.0 |
0.2 |
GO:0015695 |
organic cation transport(GO:0015695) |
| 0.0 |
0.4 |
GO:0008589 |
regulation of smoothened signaling pathway(GO:0008589) |
| 0.0 |
0.1 |
GO:0007285 |
primary spermatocyte growth(GO:0007285) |
| 0.0 |
2.7 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.1 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.0 |
4.7 |
GO:0016049 |
cell growth(GO:0016049) |
| 0.0 |
0.1 |
GO:0019441 |
tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 |
0.6 |
GO:0030520 |
intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 |
1.4 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 |
0.1 |
GO:0010172 |
embryonic body morphogenesis(GO:0010172) |
| 0.0 |
0.2 |
GO:0051924 |
regulation of calcium ion transport(GO:0051924) |
| 0.0 |
3.9 |
GO:0006457 |
protein folding(GO:0006457) |
| 0.0 |
0.8 |
GO:0006914 |
autophagy(GO:0006914) |
| 0.0 |
0.0 |
GO:0090025 |
regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 |
0.3 |
GO:0006386 |
transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 |
0.4 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.0 |
0.2 |
GO:0046676 |
negative regulation of peptide secretion(GO:0002792) negative regulation of insulin secretion(GO:0046676) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.0 |
0.1 |
GO:0045329 |
amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 |
0.2 |
GO:0021846 |
cell proliferation in forebrain(GO:0021846) |
| 0.0 |
0.5 |
GO:0042552 |
myelination(GO:0042552) |
| 0.0 |
0.3 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
0.0 |
GO:0051150 |
regulation of smooth muscle cell differentiation(GO:0051150) |
| 0.0 |
0.3 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
0.0 |
GO:0090047 |
obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 |
0.1 |
GO:0008653 |
lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 |
0.2 |
GO:0007342 |
fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 |
0.3 |
GO:0006953 |
acute-phase response(GO:0006953) |
| 0.0 |
0.4 |
GO:0006986 |
response to unfolded protein(GO:0006986) |
| 0.0 |
0.1 |
GO:0009451 |
RNA modification(GO:0009451) |
| 0.0 |
0.0 |
GO:0060423 |
foregut regionalization(GO:0060423) lung field specification(GO:0060424) primary lung bud formation(GO:0060431) lung induction(GO:0060492) regulation of branching involved in lung morphogenesis(GO:0061046) positive regulation of branching involved in lung morphogenesis(GO:0061047) iris morphogenesis(GO:0061072) |
| 0.0 |
0.2 |
GO:0015804 |
neutral amino acid transport(GO:0015804) |
| 0.0 |
0.0 |
GO:0044253 |
positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 |
0.1 |
GO:0071772 |
BMP signaling pathway(GO:0030509) response to BMP(GO:0071772) cellular response to BMP stimulus(GO:0071773) |
| 0.0 |
0.2 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.1 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 |
0.3 |
GO:0035338 |
long-chain fatty-acyl-CoA metabolic process(GO:0035336) fatty-acyl-CoA metabolic process(GO:0035337) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.0 |
0.2 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.1 |
GO:0006108 |
malate metabolic process(GO:0006108) |
| 0.0 |
0.2 |
GO:0007566 |
embryo implantation(GO:0007566) |
| 0.0 |
0.1 |
GO:0006474 |
N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 |
0.3 |
GO:0006891 |
intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 |
0.2 |
GO:0007286 |
spermatid development(GO:0007286) |
| 0.0 |
0.1 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.0 |
0.2 |
GO:0005980 |
glycogen catabolic process(GO:0005980) |
| 0.0 |
1.3 |
GO:0007507 |
heart development(GO:0007507) |
| 0.0 |
0.6 |
GO:0030521 |
androgen receptor signaling pathway(GO:0030521) |
| 0.0 |
0.0 |
GO:0009629 |
response to gravity(GO:0009629) |
| 0.0 |
0.0 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |