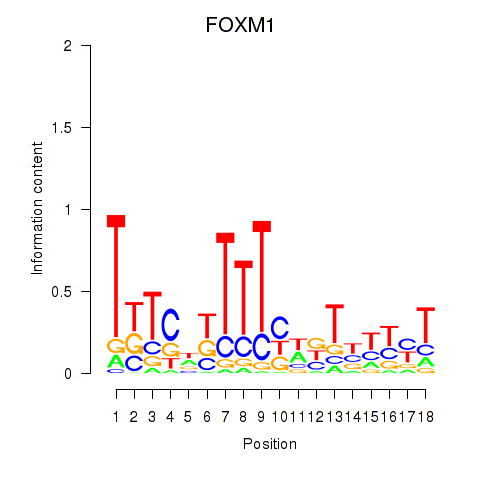
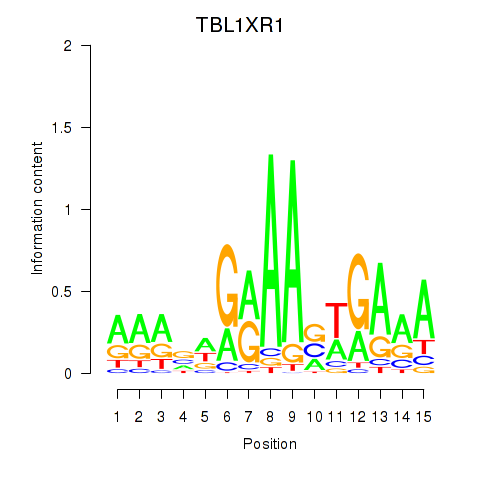
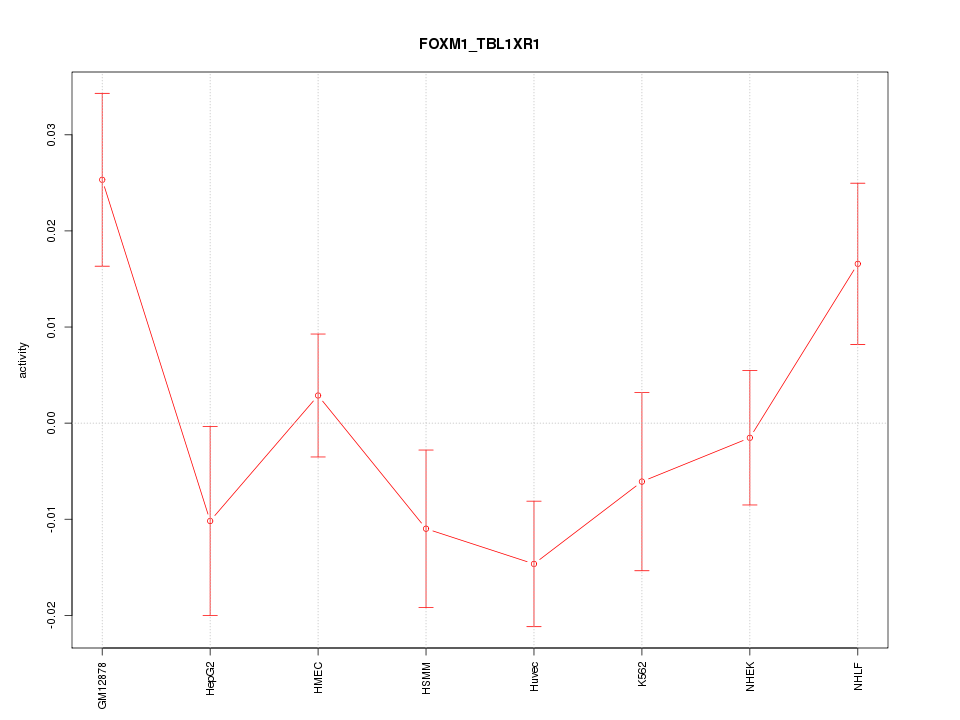
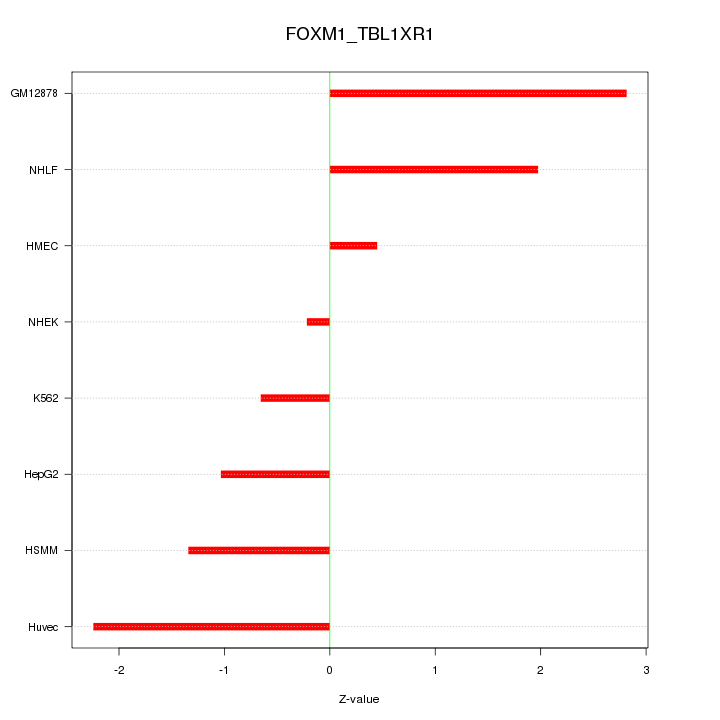
| 0.8 |
3.1 |
GO:0051712 |
positive regulation of killing of cells of other organism(GO:0051712) |
| 0.8 |
8.5 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.7 |
16.6 |
GO:0002504 |
antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.6 |
1.9 |
GO:0002678 |
positive regulation of chronic inflammatory response(GO:0002678) regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.6 |
1.8 |
GO:0002291 |
T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.5 |
1.5 |
GO:0060028 |
convergent extension involved in axis elongation(GO:0060028) |
| 0.4 |
1.6 |
GO:0031580 |
membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.4 |
2.5 |
GO:0015755 |
fructose transport(GO:0015755) |
| 0.4 |
12.7 |
GO:0050853 |
B cell receptor signaling pathway(GO:0050853) |
| 0.4 |
4.3 |
GO:1901797 |
negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.4 |
1.1 |
GO:0051097 |
negative regulation of helicase activity(GO:0051097) |
| 0.3 |
2.1 |
GO:2000124 |
cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 |
2.1 |
GO:0009441 |
glycolate metabolic process(GO:0009441) |
| 0.3 |
1.6 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.3 |
1.0 |
GO:0019441 |
tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.3 |
0.3 |
GO:0046110 |
xanthine metabolic process(GO:0046110) |
| 0.3 |
0.8 |
GO:0046586 |
regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.3 |
1.7 |
GO:0002313 |
mature B cell differentiation involved in immune response(GO:0002313) |
| 0.3 |
0.8 |
GO:0070340 |
detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.3 |
0.3 |
GO:0045937 |
positive regulation of phosphorus metabolic process(GO:0010562) positive regulation of phosphate metabolic process(GO:0045937) |
| 0.2 |
1.2 |
GO:0046952 |
ketone body catabolic process(GO:0046952) |
| 0.2 |
0.7 |
GO:0045576 |
mast cell activation(GO:0045576) |
| 0.2 |
0.2 |
GO:0045342 |
MHC class II biosynthetic process(GO:0045342) regulation of MHC class II biosynthetic process(GO:0045346) |
| 0.2 |
1.5 |
GO:0043316 |
natural killer cell activation involved in immune response(GO:0002323) cytotoxic T cell degranulation(GO:0043316) natural killer cell degranulation(GO:0043320) |
| 0.2 |
1.0 |
GO:0050965 |
detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.2 |
0.2 |
GO:0002431 |
Fc receptor mediated stimulatory signaling pathway(GO:0002431) regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.2 |
0.9 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.2 |
2.5 |
GO:0006975 |
DNA damage induced protein phosphorylation(GO:0006975) |
| 0.2 |
1.1 |
GO:0006216 |
cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.2 |
0.9 |
GO:0006848 |
pyruvate transport(GO:0006848) |
| 0.2 |
1.1 |
GO:0008588 |
release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.2 |
0.2 |
GO:0051902 |
negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.2 |
5.4 |
GO:0002474 |
antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.2 |
1.2 |
GO:0006880 |
intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 |
2.2 |
GO:0006030 |
chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 |
0.8 |
GO:0033622 |
integrin activation(GO:0033622) regulation of integrin activation(GO:0033623) positive regulation of integrin activation(GO:0033625) |
| 0.2 |
0.6 |
GO:0033632 |
regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.2 |
0.2 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.2 |
1.3 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.2 |
1.2 |
GO:2000111 |
positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.2 |
1.0 |
GO:0060100 |
regulation of phagocytosis, engulfment(GO:0060099) positive regulation of phagocytosis, engulfment(GO:0060100) regulation of membrane invagination(GO:1905153) positive regulation of membrane invagination(GO:1905155) |
| 0.2 |
0.2 |
GO:0032329 |
serine transport(GO:0032329) |
| 0.2 |
2.9 |
GO:0032814 |
regulation of natural killer cell activation(GO:0032814) |
| 0.2 |
3.0 |
GO:0070206 |
protein trimerization(GO:0070206) |
| 0.2 |
1.8 |
GO:0019987 |
obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.2 |
0.7 |
GO:0045423 |
regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.2 |
0.5 |
GO:0006549 |
isoleucine metabolic process(GO:0006549) |
| 0.2 |
0.2 |
GO:0042986 |
positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.2 |
0.6 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.2 |
0.3 |
GO:0010701 |
positive regulation of norepinephrine secretion(GO:0010701) |
| 0.2 |
0.2 |
GO:0048861 |
leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.2 |
0.2 |
GO:1902414 |
protein localization to adherens junction(GO:0071896) protein localization to cell junction(GO:1902414) |
| 0.2 |
0.5 |
GO:0002541 |
activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.2 |
0.9 |
GO:0006772 |
thiamine metabolic process(GO:0006772) |
| 0.2 |
1.2 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.2 |
0.5 |
GO:2000121 |
regulation of removal of superoxide radicals(GO:2000121) |
| 0.2 |
0.6 |
GO:0019805 |
quinolinate biosynthetic process(GO:0019805) |
| 0.1 |
0.4 |
GO:0019747 |
regulation of isoprenoid metabolic process(GO:0019747) |
| 0.1 |
0.1 |
GO:0043049 |
otic placode formation(GO:0043049) |
| 0.1 |
4.3 |
GO:0030224 |
monocyte differentiation(GO:0030224) |
| 0.1 |
0.1 |
GO:0045586 |
regulation of gamma-delta T cell differentiation(GO:0045586) positive regulation of gamma-delta T cell differentiation(GO:0045588) |
| 0.1 |
0.4 |
GO:0033364 |
mast cell secretory granule organization(GO:0033364) |
| 0.1 |
0.4 |
GO:0042088 |
T-helper 1 type immune response(GO:0042088) |
| 0.1 |
2.1 |
GO:0043330 |
response to exogenous dsRNA(GO:0043330) |
| 0.1 |
0.4 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 |
3.8 |
GO:0002286 |
T cell activation involved in immune response(GO:0002286) |
| 0.1 |
0.8 |
GO:0032897 |
negative regulation of viral transcription(GO:0032897) |
| 0.1 |
0.5 |
GO:0071418 |
cellular response to amine stimulus(GO:0071418) |
| 0.1 |
0.5 |
GO:0043653 |
mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 |
0.1 |
GO:0046874 |
quinolinate metabolic process(GO:0046874) |
| 0.1 |
0.7 |
GO:0060638 |
mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.1 |
0.4 |
GO:0043370 |
regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043370) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) regulation of CD4-positive, alpha-beta T cell activation(GO:2000514) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) |
| 0.1 |
0.1 |
GO:0045046 |
protein import into peroxisome membrane(GO:0045046) |
| 0.1 |
0.4 |
GO:0002360 |
T cell lineage commitment(GO:0002360) |
| 0.1 |
0.4 |
GO:0010224 |
response to UV-B(GO:0010224) |
| 0.1 |
0.7 |
GO:0046813 |
receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.1 |
1.0 |
GO:0007185 |
transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 |
0.8 |
GO:0033261 |
obsolete regulation of S phase(GO:0033261) |
| 0.1 |
0.2 |
GO:0006154 |
adenosine catabolic process(GO:0006154) |
| 0.1 |
0.5 |
GO:0046121 |
deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 |
0.4 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 |
0.5 |
GO:0015917 |
aminophospholipid transport(GO:0015917) |
| 0.1 |
0.1 |
GO:0046339 |
diacylglycerol biosynthetic process(GO:0006651) diacylglycerol metabolic process(GO:0046339) |
| 0.1 |
1.6 |
GO:0015939 |
pantothenate metabolic process(GO:0015939) |
| 0.1 |
0.4 |
GO:0006768 |
biotin metabolic process(GO:0006768) |
| 0.1 |
0.4 |
GO:0050968 |
detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 |
2.3 |
GO:0045730 |
respiratory burst(GO:0045730) |
| 0.1 |
0.3 |
GO:0006782 |
protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.1 |
0.3 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 |
0.3 |
GO:0046951 |
cellular ketone body metabolic process(GO:0046950) ketone body biosynthetic process(GO:0046951) |
| 0.1 |
0.3 |
GO:0071918 |
urea transmembrane transport(GO:0071918) |
| 0.1 |
0.1 |
GO:0006825 |
copper ion transport(GO:0006825) |
| 0.1 |
0.3 |
GO:0048865 |
stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.1 |
0.3 |
GO:1903961 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.1 |
1.5 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 |
1.1 |
GO:0043124 |
negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 |
0.4 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.1 |
0.6 |
GO:0007202 |
activation of phospholipase C activity(GO:0007202) positive regulation of phospholipase C activity(GO:0010863) regulation of phospholipase C activity(GO:1900274) |
| 0.1 |
0.3 |
GO:0000738 |
DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 |
0.5 |
GO:0060414 |
aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 |
0.4 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.1 |
0.2 |
GO:0050942 |
positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.1 |
0.5 |
GO:0045084 |
positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 |
0.2 |
GO:0051450 |
myoblast proliferation(GO:0051450) |
| 0.1 |
0.4 |
GO:0046642 |
negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 |
0.5 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.1 |
0.3 |
GO:0043006 |
activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.1 |
0.4 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 |
0.5 |
GO:0001661 |
conditioned taste aversion(GO:0001661) |
| 0.1 |
0.3 |
GO:0071460 |
cellular response to cell-matrix adhesion(GO:0071460) |
| 0.1 |
0.3 |
GO:0032497 |
detection of molecule of bacterial origin(GO:0032490) detection of lipopolysaccharide(GO:0032497) |
| 0.1 |
0.5 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.1 |
0.4 |
GO:0051573 |
negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.1 |
0.2 |
GO:0000733 |
DNA strand renaturation(GO:0000733) |
| 0.1 |
0.3 |
GO:0006089 |
lactate metabolic process(GO:0006089) |
| 0.1 |
0.1 |
GO:0042472 |
inner ear morphogenesis(GO:0042472) |
| 0.1 |
0.6 |
GO:0044413 |
evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 |
0.5 |
GO:0032472 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) Golgi calcium ion transport(GO:0032472) manganese ion homeostasis(GO:0055071) |
| 0.1 |
4.5 |
GO:0071260 |
cellular response to mechanical stimulus(GO:0071260) |
| 0.1 |
0.4 |
GO:0022614 |
membrane to membrane docking(GO:0022614) |
| 0.1 |
0.5 |
GO:0007499 |
ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 |
0.2 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.1 |
0.4 |
GO:0001832 |
blastocyst growth(GO:0001832) |
| 0.1 |
0.3 |
GO:0019371 |
cyclooxygenase pathway(GO:0019371) |
| 0.1 |
0.6 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 |
0.2 |
GO:0002335 |
mature B cell differentiation(GO:0002335) |
| 0.1 |
0.3 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 0.1 |
0.2 |
GO:0050857 |
positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.1 |
0.2 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 |
0.6 |
GO:0001782 |
B cell homeostasis(GO:0001782) |
| 0.1 |
0.2 |
GO:0046902 |
regulation of mitochondrial membrane permeability(GO:0046902) regulation of membrane permeability(GO:0090559) |
| 0.1 |
0.2 |
GO:0007285 |
primary spermatocyte growth(GO:0007285) |
| 0.1 |
0.4 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 |
1.9 |
GO:0000722 |
telomere maintenance via recombination(GO:0000722) |
| 0.1 |
0.1 |
GO:0097435 |
extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.1 |
0.2 |
GO:0032328 |
alanine transport(GO:0032328) |
| 0.1 |
0.2 |
GO:0030205 |
dermatan sulfate metabolic process(GO:0030205) dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 |
0.6 |
GO:0002553 |
histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 |
0.9 |
GO:0006734 |
NADH metabolic process(GO:0006734) |
| 0.1 |
0.2 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 |
1.2 |
GO:0006271 |
DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.1 |
3.2 |
GO:0030183 |
B cell differentiation(GO:0030183) |
| 0.1 |
0.7 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.1 |
0.1 |
GO:0007403 |
glial cell fate determination(GO:0007403) |
| 0.1 |
0.2 |
GO:0007079 |
mitotic chromosome movement towards spindle pole(GO:0007079) chromosome movement towards spindle pole(GO:0051305) |
| 0.1 |
0.5 |
GO:0042483 |
negative regulation of odontogenesis(GO:0042483) |
| 0.1 |
0.1 |
GO:0032510 |
endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 |
0.2 |
GO:0019242 |
methylglyoxal biosynthetic process(GO:0019242) |
| 0.1 |
1.5 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.1 |
0.2 |
GO:0002673 |
regulation of acute inflammatory response(GO:0002673) positive regulation of acute inflammatory response(GO:0002675) |
| 0.1 |
0.2 |
GO:0010836 |
negative regulation of protein ADP-ribosylation(GO:0010836) negative regulation of protein glycosylation(GO:0060051) |
| 0.1 |
0.5 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.1 |
0.1 |
GO:0010044 |
response to aluminum ion(GO:0010044) |
| 0.1 |
0.1 |
GO:0006664 |
glycolipid metabolic process(GO:0006664) |
| 0.1 |
0.1 |
GO:0048304 |
positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 |
0.4 |
GO:0006857 |
oligopeptide transport(GO:0006857) |
| 0.1 |
0.4 |
GO:0035455 |
response to interferon-alpha(GO:0035455) |
| 0.1 |
0.4 |
GO:0043402 |
glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 |
0.4 |
GO:0032527 |
retrograde protein transport, ER to cytosol(GO:0030970) protein exit from endoplasmic reticulum(GO:0032527) |
| 0.1 |
0.2 |
GO:0003351 |
epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 |
0.2 |
GO:0051255 |
spindle midzone assembly(GO:0051255) meiotic spindle midzone assembly(GO:0051257) |
| 0.1 |
0.4 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.1 |
0.3 |
GO:0000019 |
regulation of mitotic recombination(GO:0000019) |
| 0.1 |
0.2 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.1 |
0.1 |
GO:0031642 |
negative regulation of myelination(GO:0031642) |
| 0.1 |
0.1 |
GO:0097190 |
apoptotic signaling pathway(GO:0097190) |
| 0.1 |
0.1 |
GO:0006776 |
vitamin A metabolic process(GO:0006776) |
| 0.1 |
0.4 |
GO:0051323 |
metaphase(GO:0051323) |
| 0.1 |
2.7 |
GO:0050690 |
regulation of defense response to virus by virus(GO:0050690) |
| 0.1 |
0.5 |
GO:0048712 |
negative regulation of astrocyte differentiation(GO:0048712) |
| 0.1 |
0.2 |
GO:0060292 |
long term synaptic depression(GO:0060292) |
| 0.1 |
0.4 |
GO:0006658 |
phosphatidylserine metabolic process(GO:0006658) |
| 0.1 |
0.1 |
GO:0001514 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 |
0.3 |
GO:0070584 |
mitochondrion morphogenesis(GO:0070584) |
| 0.1 |
0.6 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.1 |
0.2 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 |
0.6 |
GO:1904031 |
positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 |
0.4 |
GO:0001837 |
epithelial to mesenchymal transition(GO:0001837) |
| 0.1 |
0.6 |
GO:0046825 |
regulation of protein export from nucleus(GO:0046825) |
| 0.1 |
4.2 |
GO:0060333 |
interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.1 |
0.3 |
GO:0010447 |
response to acidic pH(GO:0010447) |
| 0.1 |
0.1 |
GO:0021836 |
cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.1 |
0.1 |
GO:0014834 |
skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.1 |
0.1 |
GO:0043149 |
contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 |
0.5 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.1 |
0.2 |
GO:0050798 |
activated T cell proliferation(GO:0050798) |
| 0.1 |
0.4 |
GO:0009136 |
ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.1 |
1.0 |
GO:0050930 |
induction of positive chemotaxis(GO:0050930) |
| 0.1 |
0.2 |
GO:0046495 |
nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 |
0.1 |
GO:0042573 |
retinoic acid metabolic process(GO:0042573) |
| 0.1 |
0.3 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.1 |
0.2 |
GO:0033688 |
regulation of osteoblast proliferation(GO:0033688) |
| 0.1 |
0.4 |
GO:0034465 |
cellular potassium ion homeostasis(GO:0030007) response to carbon monoxide(GO:0034465) negative regulation of cell volume(GO:0045794) |
| 0.1 |
0.3 |
GO:1902603 |
carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.1 |
0.2 |
GO:0032494 |
response to peptidoglycan(GO:0032494) |
| 0.1 |
0.2 |
GO:2001259 |
positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of cation channel activity(GO:2001259) |
| 0.1 |
0.1 |
GO:0051790 |
short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 |
0.4 |
GO:0006477 |
protein sulfation(GO:0006477) |
| 0.1 |
0.2 |
GO:0051301 |
cell division(GO:0051301) |
| 0.1 |
0.5 |
GO:0045898 |
regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 |
0.1 |
GO:0055015 |
ventricular cardiac muscle cell development(GO:0055015) |
| 0.1 |
0.2 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 |
0.2 |
GO:0007215 |
glutamate receptor signaling pathway(GO:0007215) |
| 0.1 |
0.4 |
GO:0006188 |
IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.1 |
0.2 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 |
0.2 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 |
0.1 |
GO:0007613 |
memory(GO:0007613) |
| 0.1 |
0.3 |
GO:0031638 |
zymogen activation(GO:0031638) |
| 0.1 |
0.2 |
GO:0010759 |
regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) |
| 0.1 |
0.2 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 |
1.0 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.1 |
0.2 |
GO:0002689 |
negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.1 |
0.2 |
GO:0051231 |
mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) |
| 0.1 |
0.2 |
GO:0009106 |
lipoate metabolic process(GO:0009106) |
| 0.1 |
0.1 |
GO:0031133 |
regulation of axon diameter(GO:0031133) regulation of cell projection size(GO:0032536) |
| 0.1 |
0.1 |
GO:0034339 |
obsolete regulation of transcription from RNA polymerase II promoter by nuclear hormone receptor(GO:0034339) |
| 0.1 |
0.3 |
GO:0043116 |
negative regulation of vascular permeability(GO:0043116) |
| 0.1 |
0.8 |
GO:1901661 |
quinone metabolic process(GO:1901661) |
| 0.1 |
0.3 |
GO:0045008 |
depyrimidination(GO:0045008) |
| 0.1 |
0.2 |
GO:0007207 |
phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 |
0.4 |
GO:0009820 |
alkaloid metabolic process(GO:0009820) |
| 0.1 |
0.4 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.1 |
1.9 |
GO:0006903 |
vesicle targeting(GO:0006903) |
| 0.0 |
0.1 |
GO:0007588 |
excretion(GO:0007588) |
| 0.0 |
1.3 |
GO:0006298 |
mismatch repair(GO:0006298) |
| 0.0 |
0.3 |
GO:0042149 |
cellular response to glucose starvation(GO:0042149) |
| 0.0 |
0.2 |
GO:0046826 |
negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 |
0.0 |
GO:0032094 |
response to food(GO:0032094) |
| 0.0 |
0.1 |
GO:0048627 |
myoblast development(GO:0048627) |
| 0.0 |
0.2 |
GO:0033147 |
negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 |
0.4 |
GO:0035590 |
purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 |
0.3 |
GO:0051290 |
protein heterotetramerization(GO:0051290) |
| 0.0 |
0.2 |
GO:0006720 |
isoprenoid metabolic process(GO:0006720) |
| 0.0 |
0.2 |
GO:0018352 |
protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 |
0.5 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.0 |
0.1 |
GO:0006382 |
adenosine to inosine editing(GO:0006382) |
| 0.0 |
0.0 |
GO:0046323 |
glucose import(GO:0046323) |
| 0.0 |
0.2 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 |
0.2 |
GO:0009649 |
entrainment of circadian clock(GO:0009649) |
| 0.0 |
2.5 |
GO:0032480 |
negative regulation of type I interferon production(GO:0032480) |
| 0.0 |
0.1 |
GO:0006051 |
mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 |
0.2 |
GO:0030225 |
macrophage differentiation(GO:0030225) |
| 0.0 |
1.9 |
GO:0019221 |
cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 |
0.2 |
GO:0060528 |
secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 |
0.2 |
GO:0010993 |
ubiquitin homeostasis(GO:0010992) regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 |
0.2 |
GO:0019255 |
glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 |
0.1 |
GO:0021527 |
spinal cord association neuron differentiation(GO:0021527) |
| 0.0 |
0.3 |
GO:0006011 |
UDP-glucose metabolic process(GO:0006011) UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 |
0.4 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 |
0.2 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 |
0.2 |
GO:0032236 |
obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 |
0.1 |
GO:0046080 |
dUTP metabolic process(GO:0046080) |
| 0.0 |
0.1 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) cytoplasmic translation(GO:0002181) cytoplasmic translational initiation(GO:0002183) |
| 0.0 |
0.1 |
GO:0010792 |
DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 |
0.1 |
GO:0045815 |
positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.0 |
0.4 |
GO:0001906 |
cell killing(GO:0001906) |
| 0.0 |
1.0 |
GO:1904894 |
positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 |
0.0 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 |
1.0 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.0 |
0.3 |
GO:0090280 |
positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) positive regulation of calcium ion import(GO:0090280) |
| 0.0 |
0.1 |
GO:0046685 |
response to arsenic-containing substance(GO:0046685) |
| 0.0 |
0.3 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 |
0.1 |
GO:1901724 |
positive regulation of cell proliferation involved in kidney development(GO:1901724) |
| 0.0 |
0.4 |
GO:0060216 |
definitive hemopoiesis(GO:0060216) |
| 0.0 |
0.4 |
GO:0010042 |
response to manganese ion(GO:0010042) |
| 0.0 |
0.6 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.0 |
0.3 |
GO:0007143 |
female meiotic division(GO:0007143) |
| 0.0 |
3.5 |
GO:0006977 |
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.0 |
0.1 |
GO:0046469 |
platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 |
0.0 |
GO:0060487 |
lung cell differentiation(GO:0060479) lung epithelial cell differentiation(GO:0060487) |
| 0.0 |
0.3 |
GO:0032516 |
positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 |
0.3 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) |
| 0.0 |
0.1 |
GO:0046579 |
positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 |
0.1 |
GO:0007060 |
male meiosis chromosome segregation(GO:0007060) |
| 0.0 |
0.5 |
GO:0016540 |
protein autoprocessing(GO:0016540) |
| 0.0 |
0.3 |
GO:0046638 |
regulation of alpha-beta T cell differentiation(GO:0046637) positive regulation of alpha-beta T cell differentiation(GO:0046638) |
| 0.0 |
0.3 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) vagina development(GO:0060068) |
| 0.0 |
0.1 |
GO:0001552 |
ovarian follicle atresia(GO:0001552) |
| 0.0 |
0.1 |
GO:0006307 |
DNA dealkylation involved in DNA repair(GO:0006307) DNA dealkylation(GO:0035510) |
| 0.0 |
0.1 |
GO:0010716 |
regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 |
0.3 |
GO:0071354 |
cellular response to interleukin-6(GO:0071354) |
| 0.0 |
0.2 |
GO:0007063 |
regulation of sister chromatid cohesion(GO:0007063) |
| 0.0 |
0.0 |
GO:0045060 |
negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 |
0.2 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.0 |
0.2 |
GO:0006290 |
pyrimidine dimer repair(GO:0006290) |
| 0.0 |
0.3 |
GO:0060122 |
auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 |
0.6 |
GO:0002639 |
positive regulation of immunoglobulin production(GO:0002639) |
| 0.0 |
0.1 |
GO:0000189 |
MAPK import into nucleus(GO:0000189) |
| 0.0 |
0.2 |
GO:0002430 |
complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 |
0.2 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 |
0.1 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.0 |
0.2 |
GO:0099515 |
nuclear migration along microfilament(GO:0031022) actin filament-based transport(GO:0099515) |
| 0.0 |
0.1 |
GO:0031999 |
negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.0 |
0.1 |
GO:0042320 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) |
| 0.0 |
0.1 |
GO:0006828 |
manganese ion transport(GO:0006828) |
| 0.0 |
0.4 |
GO:0030101 |
natural killer cell activation(GO:0030101) |
| 0.0 |
0.1 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.0 |
0.1 |
GO:0034723 |
DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 |
2.2 |
GO:0031295 |
lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 |
0.3 |
GO:0006600 |
creatine metabolic process(GO:0006600) |
| 0.0 |
0.1 |
GO:0045085 |
negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 |
0.2 |
GO:0060760 |
positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.0 |
0.2 |
GO:0060676 |
ureteric bud formation(GO:0060676) mesonephric tubule formation(GO:0072172) |
| 0.0 |
0.6 |
GO:0000303 |
response to superoxide(GO:0000303) |
| 0.0 |
0.7 |
GO:0007032 |
endosome organization(GO:0007032) |
| 0.0 |
0.1 |
GO:0050883 |
musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 |
0.1 |
GO:0033578 |
protein glycosylation in Golgi(GO:0033578) |
| 0.0 |
0.1 |
GO:0006573 |
valine metabolic process(GO:0006573) |
| 0.0 |
0.1 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.1 |
GO:0071848 |
regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) TNFSF11-mediated signaling pathway(GO:0071847) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.0 |
0.1 |
GO:0060049 |
regulation of protein ADP-ribosylation(GO:0010835) nitric oxide homeostasis(GO:0033484) positive regulation of synapse assembly(GO:0051965) regulation of protein glycosylation(GO:0060049) |
| 0.0 |
0.1 |
GO:0031033 |
myosin filament organization(GO:0031033) |
| 0.0 |
0.1 |
GO:0048859 |
formation of anatomical boundary(GO:0048859) |
| 0.0 |
0.5 |
GO:0042113 |
B cell activation(GO:0042113) |
| 0.0 |
0.1 |
GO:0046851 |
negative regulation of bone resorption(GO:0045779) negative regulation of bone remodeling(GO:0046851) |
| 0.0 |
0.3 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
0.1 |
GO:0051823 |
regulation of synapse structural plasticity(GO:0051823) |
| 0.0 |
0.1 |
GO:0035283 |
central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 |
0.1 |
GO:0042226 |
interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 |
0.1 |
GO:0019059 |
obsolete initiation of viral infection(GO:0019059) |
| 0.0 |
0.3 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.0 |
0.1 |
GO:0003056 |
regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 |
0.1 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
1.5 |
GO:0031397 |
negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 |
0.2 |
GO:0040015 |
negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 |
0.1 |
GO:0007398 |
ectoderm development(GO:0007398) |
| 0.0 |
0.1 |
GO:0003032 |
detection of oxygen(GO:0003032) |
| 0.0 |
0.1 |
GO:0016265 |
obsolete death(GO:0016265) |
| 0.0 |
0.0 |
GO:0002544 |
chronic inflammatory response(GO:0002544) |
| 0.0 |
0.5 |
GO:0006449 |
regulation of translational termination(GO:0006449) |
| 0.0 |
0.1 |
GO:0051968 |
positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 |
0.0 |
GO:0030432 |
peristalsis(GO:0030432) |
| 0.0 |
0.1 |
GO:0002507 |
tolerance induction(GO:0002507) |
| 0.0 |
0.1 |
GO:0006435 |
threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 |
0.2 |
GO:0051017 |
actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.0 |
0.2 |
GO:0035282 |
segmentation(GO:0035282) |
| 0.0 |
2.6 |
GO:0006338 |
chromatin remodeling(GO:0006338) |
| 0.0 |
0.0 |
GO:0009247 |
glycolipid biosynthetic process(GO:0009247) |
| 0.0 |
0.1 |
GO:0060291 |
long-term synaptic potentiation(GO:0060291) |
| 0.0 |
0.2 |
GO:0000012 |
single strand break repair(GO:0000012) |
| 0.0 |
0.1 |
GO:0002281 |
macrophage activation involved in immune response(GO:0002281) |
| 0.0 |
0.4 |
GO:0032876 |
regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 |
0.0 |
GO:0030948 |
negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 |
0.3 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.0 |
0.2 |
GO:0044245 |
polysaccharide digestion(GO:0044245) |
| 0.0 |
0.1 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.0 |
0.0 |
GO:0006542 |
glutamine biosynthetic process(GO:0006542) |
| 0.0 |
0.2 |
GO:0003416 |
endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.0 |
0.1 |
GO:0046546 |
male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 |
0.1 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.0 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 |
0.1 |
GO:0021938 |
smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 |
0.7 |
GO:0034112 |
positive regulation of homotypic cell-cell adhesion(GO:0034112) |
| 0.0 |
0.1 |
GO:0010157 |
response to chlorate(GO:0010157) |
| 0.0 |
0.3 |
GO:0033209 |
tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 |
0.1 |
GO:0033002 |
muscle cell proliferation(GO:0033002) |
| 0.0 |
0.1 |
GO:0006669 |
sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 |
0.1 |
GO:0018342 |
protein prenylation(GO:0018342) protein geranylgeranylation(GO:0018344) prenylation(GO:0097354) |
| 0.0 |
0.3 |
GO:0023058 |
desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) adaptation of signaling pathway(GO:0023058) |
| 0.0 |
0.1 |
GO:0000394 |
RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 |
0.4 |
GO:0046676 |
negative regulation of insulin secretion(GO:0046676) |
| 0.0 |
0.1 |
GO:0034625 |
fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 |
0.0 |
GO:0042481 |
regulation of odontogenesis(GO:0042481) |
| 0.0 |
0.2 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 |
1.7 |
GO:0051092 |
positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 |
0.0 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 |
0.2 |
GO:0001561 |
fatty acid alpha-oxidation(GO:0001561) |
| 0.0 |
0.7 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 |
0.0 |
GO:0034614 |
cellular response to reactive oxygen species(GO:0034614) |
| 0.0 |
0.1 |
GO:0051123 |
RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 |
0.1 |
GO:0019471 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 |
0.1 |
GO:0046015 |
carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.0 |
0.1 |
GO:0046185 |
aldehyde catabolic process(GO:0046185) |
| 0.0 |
0.0 |
GO:0008333 |
endosome to lysosome transport(GO:0008333) |
| 0.0 |
0.1 |
GO:0032506 |
cytokinetic process(GO:0032506) |
| 0.0 |
0.1 |
GO:0035120 |
post-embryonic appendage morphogenesis(GO:0035120) |
| 0.0 |
0.1 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 |
0.7 |
GO:0051225 |
spindle assembly(GO:0051225) |
| 0.0 |
0.1 |
GO:0060113 |
inner ear receptor cell differentiation(GO:0060113) |
| 0.0 |
0.1 |
GO:0001831 |
trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 |
0.1 |
GO:0006295 |
nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 |
0.1 |
GO:0051877 |
pigment granule aggregation in cell center(GO:0051877) |
| 0.0 |
0.1 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.0 |
0.1 |
GO:1904396 |
regulation of synaptic growth at neuromuscular junction(GO:0008582) regulation of neuromuscular junction development(GO:1904396) |
| 0.0 |
0.1 |
GO:0016046 |
detection of fungus(GO:0016046) |
| 0.0 |
0.1 |
GO:0072078 |
ureteric bud morphogenesis(GO:0060675) renal tubule morphogenesis(GO:0061333) nephron tubule morphogenesis(GO:0072078) mesonephric tubule morphogenesis(GO:0072171) |
| 0.0 |
0.1 |
GO:0060649 |
nipple development(GO:0060618) mammary gland bud elongation(GO:0060649) nipple morphogenesis(GO:0060658) nipple sheath formation(GO:0060659) |
| 0.0 |
0.1 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.0 |
0.1 |
GO:0060014 |
granulosa cell differentiation(GO:0060014) |
| 0.0 |
0.1 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 0.0 |
0.1 |
GO:0071480 |
cellular response to gamma radiation(GO:0071480) |
| 0.0 |
0.2 |
GO:0015781 |
pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 |
0.2 |
GO:0071044 |
histone mRNA catabolic process(GO:0071044) |
| 0.0 |
0.1 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.0 |
0.1 |
GO:0045767 |
obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 |
0.1 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 |
0.2 |
GO:0046835 |
carbohydrate phosphorylation(GO:0046835) |
| 0.0 |
0.1 |
GO:0035121 |
obsolete tail morphogenesis(GO:0035121) |
| 0.0 |
0.1 |
GO:0021898 |
commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 |
0.1 |
GO:0006780 |
uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.0 |
0.1 |
GO:0060347 |
heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 |
1.7 |
GO:0009615 |
response to virus(GO:0009615) |
| 0.0 |
0.1 |
GO:0006829 |
zinc II ion transport(GO:0006829) |
| 0.0 |
0.2 |
GO:0032964 |
collagen biosynthetic process(GO:0032964) |
| 0.0 |
0.0 |
GO:0051083 |
'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 |
0.1 |
GO:0035608 |
protein deglutamylation(GO:0035608) |
| 0.0 |
0.1 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.0 |
0.8 |
GO:0000080 |
mitotic G1 phase(GO:0000080) |
| 0.0 |
0.1 |
GO:0045086 |
positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 |
0.1 |
GO:0045880 |
positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 |
0.1 |
GO:0070493 |
thrombin receptor signaling pathway(GO:0070493) |
| 0.0 |
0.2 |
GO:0009118 |
regulation of nucleoside metabolic process(GO:0009118) regulation of ATP metabolic process(GO:1903578) |
| 0.0 |
0.0 |
GO:0021675 |
nerve development(GO:0021675) |
| 0.0 |
0.2 |
GO:0046618 |
drug export(GO:0046618) |
| 0.0 |
0.1 |
GO:0016074 |
snoRNA metabolic process(GO:0016074) |
| 0.0 |
0.2 |
GO:0046831 |
regulation of RNA export from nucleus(GO:0046831) |
| 0.0 |
0.1 |
GO:0016476 |
regulation of embryonic cell shape(GO:0016476) |
| 0.0 |
0.2 |
GO:0051930 |
regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 |
0.0 |
GO:0032413 |
negative regulation of ion transmembrane transporter activity(GO:0032413) negative regulation of cation transmembrane transport(GO:1904063) |
| 0.0 |
0.1 |
GO:0043114 |
regulation of vascular permeability(GO:0043114) |
| 0.0 |
0.1 |
GO:0072010 |
renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 |
0.3 |
GO:0048536 |
spleen development(GO:0048536) |
| 0.0 |
0.1 |
GO:0016045 |
detection of bacterium(GO:0016045) |
| 0.0 |
0.1 |
GO:0000090 |
mitotic anaphase(GO:0000090) |
| 0.0 |
0.0 |
GO:0010159 |
specification of organ position(GO:0010159) |
| 0.0 |
0.1 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.1 |
GO:0050907 |
detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 |
0.0 |
GO:0030578 |
nuclear body organization(GO:0030575) PML body organization(GO:0030578) |
| 0.0 |
0.4 |
GO:0032456 |
endocytic recycling(GO:0032456) |
| 0.0 |
0.1 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.0 |
GO:0052572 |
response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.0 |
0.1 |
GO:2000178 |
negative regulation of neuroblast proliferation(GO:0007406) forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 |
0.1 |
GO:0009048 |
dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 |
0.2 |
GO:0048240 |
sperm capacitation(GO:0048240) |
| 0.0 |
0.3 |
GO:0008347 |
glial cell migration(GO:0008347) |
| 0.0 |
0.1 |
GO:0060529 |
squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.0 |
0.6 |
GO:0000723 |
telomere maintenance(GO:0000723) |
| 0.0 |
0.2 |
GO:0032836 |
glomerular basement membrane development(GO:0032836) |
| 0.0 |
0.1 |
GO:0003084 |
positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 |
0.0 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.0 |
0.0 |
GO:0014045 |
establishment of endothelial blood-brain barrier(GO:0014045) central nervous system vasculogenesis(GO:0022009) |
| 0.0 |
0.2 |
GO:0043525 |
positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.0 |
0.2 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 |
0.0 |
GO:0060670 |
branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 |
0.2 |
GO:0042993 |
positive regulation of transcription factor import into nucleus(GO:0042993) |
| 0.0 |
0.0 |
GO:0007140 |
male meiosis(GO:0007140) |
| 0.0 |
0.1 |
GO:0043095 |
regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 |
0.8 |
GO:0006626 |
protein targeting to mitochondrion(GO:0006626) |
| 0.0 |
0.2 |
GO:0045672 |
positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 |
0.1 |
GO:0060080 |
synaptic transmission, glycinergic(GO:0060012) inhibitory postsynaptic potential(GO:0060080) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 |
0.0 |
GO:0006104 |
succinyl-CoA metabolic process(GO:0006104) |
| 0.0 |
0.0 |
GO:0017085 |
response to insecticide(GO:0017085) |
| 0.0 |
0.0 |
GO:0043278 |
response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 |
0.2 |
GO:0046839 |
phospholipid dephosphorylation(GO:0046839) |
| 0.0 |
0.6 |
GO:0050931 |
melanocyte differentiation(GO:0030318) pigment cell differentiation(GO:0050931) |
| 0.0 |
0.4 |
GO:0000216 |
obsolete M/G1 transition of mitotic cell cycle(GO:0000216) |
| 0.0 |
0.1 |
GO:0006105 |
succinate metabolic process(GO:0006105) |
| 0.0 |
0.0 |
GO:0030825 |
positive regulation of cGMP metabolic process(GO:0030825) positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 |
0.3 |
GO:0030321 |
transepithelial chloride transport(GO:0030321) |
| 0.0 |
0.1 |
GO:0051588 |
regulation of neurotransmitter transport(GO:0051588) |
| 0.0 |
0.2 |
GO:0051647 |
nucleus localization(GO:0051647) |
| 0.0 |
0.0 |
GO:0048755 |
branching morphogenesis of a nerve(GO:0048755) |
| 0.0 |
0.1 |
GO:0018916 |
nitrobenzene metabolic process(GO:0018916) |
| 0.0 |
0.2 |
GO:0006839 |
mitochondrial transport(GO:0006839) |
| 0.0 |
0.2 |
GO:0006546 |
glycine catabolic process(GO:0006546) |
| 0.0 |
0.1 |
GO:0033683 |
nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 |
0.0 |
GO:0090047 |
retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 |
0.3 |
GO:0030838 |
positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 |
0.9 |
GO:0007193 |
adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 |
0.1 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 |
0.1 |
GO:0048853 |
forebrain morphogenesis(GO:0048853) |
| 0.0 |
0.6 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
1.2 |
GO:0051291 |
protein heterooligomerization(GO:0051291) |
| 0.0 |
0.1 |
GO:0014046 |
dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 |
0.1 |
GO:0060644 |
mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 |
0.1 |
GO:0060456 |
positive regulation of digestive system process(GO:0060456) |
| 0.0 |
0.0 |
GO:0018283 |
metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 |
0.1 |
GO:0015802 |
basic amino acid transport(GO:0015802) |
| 0.0 |
0.0 |
GO:0060054 |
positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 |
0.1 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 |
0.1 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.3 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.0 |
0.1 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.0 |
0.3 |
GO:0032024 |
positive regulation of insulin secretion(GO:0032024) |
| 0.0 |
0.0 |
GO:0002053 |
positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 |
0.1 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 |
0.2 |
GO:0006704 |
glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 |
0.1 |
GO:0051249 |
regulation of lymphocyte activation(GO:0051249) |
| 0.0 |
0.1 |
GO:0046349 |
UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.0 |
0.0 |
GO:0046487 |
glyoxylate metabolic process(GO:0046487) |
| 0.0 |
0.1 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 |
0.1 |
GO:0014049 |
positive regulation of glutamate secretion(GO:0014049) |
| 0.0 |
0.1 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 |
0.3 |
GO:0045069 |
regulation of viral genome replication(GO:0045069) |
| 0.0 |
0.2 |
GO:0071230 |
cellular response to amino acid stimulus(GO:0071230) |
| 0.0 |
0.1 |
GO:0007028 |
cytoplasm organization(GO:0007028) |
| 0.0 |
0.1 |
GO:0006264 |
mitochondrial DNA replication(GO:0006264) |
| 0.0 |
0.0 |
GO:0060259 |
regulation of feeding behavior(GO:0060259) |
| 0.0 |
0.1 |
GO:0043550 |
regulation of lipid kinase activity(GO:0043550) |
| 0.0 |
0.1 |
GO:0021680 |
cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 |
0.7 |
GO:0051289 |
protein homotetramerization(GO:0051289) |
| 0.0 |
0.0 |
GO:0030168 |
platelet activation(GO:0030168) |
| 0.0 |
0.1 |
GO:0009313 |
oligosaccharide catabolic process(GO:0009313) |
| 0.0 |
0.0 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 |
0.0 |
GO:0000729 |
DNA double-strand break processing(GO:0000729) |
| 0.0 |
0.1 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
0.2 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.0 |
0.1 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 |
0.1 |
GO:0001946 |
lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 0.0 |
0.1 |
GO:0034036 |
purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 |
0.3 |
GO:0018298 |
protein-chromophore linkage(GO:0018298) |
| 0.0 |
0.1 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 |
1.2 |
GO:0000236 |
mitotic prometaphase(GO:0000236) |
| 0.0 |
0.1 |
GO:0000059 |
protein import into nucleus, docking(GO:0000059) |
| 0.0 |
0.0 |
GO:0051852 |
disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 |
0.0 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) female courtship behavior(GO:0008050) regulation of female receptivity(GO:0045924) female mating behavior(GO:0060180) |
| 0.0 |
0.0 |
GO:0009067 |
aspartate family amino acid biosynthetic process(GO:0009067) |
| 0.0 |
0.6 |
GO:0006888 |
ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 |
0.1 |
GO:0006655 |
phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 |
0.5 |
GO:0046651 |
lymphocyte proliferation(GO:0046651) |
| 0.0 |
0.0 |
GO:0007626 |
locomotory behavior(GO:0007626) |
| 0.0 |
0.1 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 |
0.1 |
GO:0032099 |
negative regulation of appetite(GO:0032099) |
| 0.0 |
0.0 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 |
0.3 |
GO:0007030 |
Golgi organization(GO:0007030) |
| 0.0 |
0.1 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.0 |
0.1 |
GO:0070189 |
kynurenine metabolic process(GO:0070189) |
| 0.0 |
0.1 |
GO:0001938 |
positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 |
0.2 |
GO:0006099 |
tricarboxylic acid cycle(GO:0006099) |
| 0.0 |
0.1 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
0.1 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 |
0.0 |
GO:0009051 |
pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 |
0.1 |
GO:0006422 |
aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 |
0.2 |
GO:0016925 |
protein sumoylation(GO:0016925) |
| 0.0 |
0.0 |
GO:0031290 |
retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 |
0.1 |
GO:0034643 |
establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 |
0.1 |
GO:0010591 |
regulation of lamellipodium assembly(GO:0010591) |
| 0.0 |
0.0 |
GO:0009791 |
post-embryonic development(GO:0009791) |
| 0.0 |
0.2 |
GO:0051131 |
chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 |
0.1 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.2 |
GO:0016558 |
protein import into peroxisome matrix(GO:0016558) |
| 0.0 |
0.0 |
GO:0006707 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.1 |
GO:0042769 |
DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 |
0.3 |
GO:0019079 |
viral genome replication(GO:0019079) |
| 0.0 |
0.0 |
GO:0040020 |
regulation of meiotic nuclear division(GO:0040020) |
| 0.0 |
0.0 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.0 |
0.0 |
GO:0031427 |
response to methotrexate(GO:0031427) |
| 0.0 |
0.1 |
GO:0045176 |
apical protein localization(GO:0045176) |
| 0.0 |
0.3 |
GO:0042136 |
neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 |
0.0 |
GO:0035112 |
genitalia morphogenesis(GO:0035112) |
| 0.0 |
0.3 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.0 |
GO:0032096 |
negative regulation of response to food(GO:0032096) |
| 0.0 |
0.0 |
GO:0009134 |
nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 |
0.6 |
GO:0050851 |
antigen receptor-mediated signaling pathway(GO:0050851) |
| 0.0 |
0.1 |
GO:0001539 |
cilium or flagellum-dependent cell motility(GO:0001539) |
| 0.0 |
0.1 |
GO:0010830 |
regulation of myotube differentiation(GO:0010830) |
| 0.0 |
0.0 |
GO:0034612 |
response to tumor necrosis factor(GO:0034612) |
| 0.0 |
0.1 |
GO:0060512 |
prostate gland morphogenesis(GO:0060512) |
| 0.0 |
0.0 |
GO:1904036 |
negative regulation of epithelial cell apoptotic process(GO:1904036) negative regulation of endothelial cell apoptotic process(GO:2000352) |
| 0.0 |
2.2 |
GO:0006184 |
obsolete GTP catabolic process(GO:0006184) |
| 0.0 |
1.6 |
GO:0007565 |
female pregnancy(GO:0007565) |
| 0.0 |
0.0 |
GO:0007617 |
mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 |
0.1 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.0 |
0.0 |
GO:0007062 |
sister chromatid cohesion(GO:0007062) |
| 0.0 |
0.2 |
GO:0006386 |
transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 |
0.1 |
GO:0032098 |
regulation of appetite(GO:0032098) |
| 0.0 |
0.1 |
GO:0045744 |
negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 |
0.0 |
GO:0006167 |
AMP biosynthetic process(GO:0006167) |
| 0.0 |
0.1 |
GO:0036260 |
7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.0 |
1.5 |
GO:0007050 |
cell cycle arrest(GO:0007050) |
| 0.0 |
0.7 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.0 |
0.2 |
GO:0000380 |
alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 |
0.2 |
GO:0000060 |
protein import into nucleus, translocation(GO:0000060) |
| 0.0 |
0.0 |
GO:0051927 |
obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.0 |
0.4 |
GO:0006309 |
DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.0 |
0.0 |
GO:0042117 |
monocyte activation(GO:0042117) PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 |
0.0 |
GO:0015959 |
diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 |
0.4 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.1 |
GO:0090080 |
positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 |
0.0 |
GO:0000098 |
sulfur amino acid catabolic process(GO:0000098) |
| 0.0 |
0.0 |
GO:0009162 |
deoxyribonucleoside monophosphate metabolic process(GO:0009162) pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) dUMP metabolic process(GO:0046078) |
| 0.0 |
0.1 |
GO:0033962 |
cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 |
0.1 |
GO:0008295 |
spermidine biosynthetic process(GO:0008295) |
| 0.0 |
0.1 |
GO:0009411 |
response to UV(GO:0009411) |
| 0.0 |
0.1 |
GO:0009396 |
folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 |
0.0 |
GO:0060976 |
coronary vasculature development(GO:0060976) |
| 0.0 |
0.0 |
GO:0001701 |
in utero embryonic development(GO:0001701) |
| 0.0 |
0.7 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.0 |
GO:0042398 |
cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.0 |
0.0 |
GO:0019919 |
peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 |
0.2 |
GO:0042438 |
melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 |
0.0 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.0 |
0.1 |
GO:0006642 |
triglyceride mobilization(GO:0006642) |
| 0.0 |
0.6 |
GO:0046777 |
protein autophosphorylation(GO:0046777) |
| 0.0 |
0.2 |
GO:0009083 |
branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
0.0 |
GO:0000076 |
DNA replication checkpoint(GO:0000076) |
| 0.0 |
0.2 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
0.0 |
GO:0032410 |
negative regulation of transporter activity(GO:0032410) |
| 0.0 |
0.1 |
GO:0006700 |
C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 |
0.0 |
GO:0003018 |
vascular process in circulatory system(GO:0003018) regulation of tube size(GO:0035150) regulation of blood vessel size(GO:0050880) |
| 0.0 |
0.1 |
GO:0001954 |
positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 |
0.0 |
GO:0071294 |
cellular response to zinc ion(GO:0071294) |
| 0.0 |
0.3 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.0 |
0.2 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.0 |
0.0 |
GO:0033363 |
secretory granule organization(GO:0033363) |
| 0.0 |
0.0 |
GO:0051196 |
regulation of cofactor metabolic process(GO:0051193) regulation of coenzyme metabolic process(GO:0051196) |
| 0.0 |
0.0 |
GO:0030261 |
chromosome condensation(GO:0030261) |
| 0.0 |
0.1 |
GO:0006301 |
postreplication repair(GO:0006301) |
| 0.0 |
0.0 |
GO:0071109 |
thalamus development(GO:0021794) superior temporal gyrus development(GO:0071109) |
| 0.0 |
0.1 |
GO:0018106 |
peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 |
0.0 |
GO:0007389 |
pattern specification process(GO:0007389) |
| 0.0 |
6.4 |
GO:0006955 |
immune response(GO:0006955) |
| 0.0 |
0.1 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.0 |
GO:0045338 |
farnesyl diphosphate metabolic process(GO:0045338) |
| 0.0 |
0.0 |
GO:1900006 |
positive regulation of dendrite development(GO:1900006) |
| 0.0 |
0.2 |
GO:0000725 |
double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 |
0.1 |
GO:0045540 |
regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 |
0.1 |
GO:0006004 |
fucose metabolic process(GO:0006004) |
| 0.0 |
0.0 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) |
| 0.0 |
0.4 |
GO:0006289 |
nucleotide-excision repair(GO:0006289) |
| 0.0 |
1.0 |
GO:0006401 |
RNA catabolic process(GO:0006401) |
| 0.0 |
0.0 |
GO:0060438 |
trachea development(GO:0060438) |
| 0.0 |
0.0 |
GO:0032924 |
activin receptor signaling pathway(GO:0032924) |
| 0.0 |
0.0 |
GO:0032613 |
interleukin-10 production(GO:0032613) |
| 0.0 |
0.0 |
GO:0016073 |
snRNA metabolic process(GO:0016073) |
| 0.0 |
0.0 |
GO:0001660 |
fever generation(GO:0001660) |
| 0.0 |
0.0 |
GO:0070254 |
mucus secretion(GO:0070254) |
| 0.0 |
0.0 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.0 |
0.0 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
0.0 |
GO:0043536 |
positive regulation of blood vessel endothelial cell migration(GO:0043536) |
| 0.0 |
0.0 |
GO:0034770 |
histone H4-K20 methylation(GO:0034770) |
| 0.0 |
0.0 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |