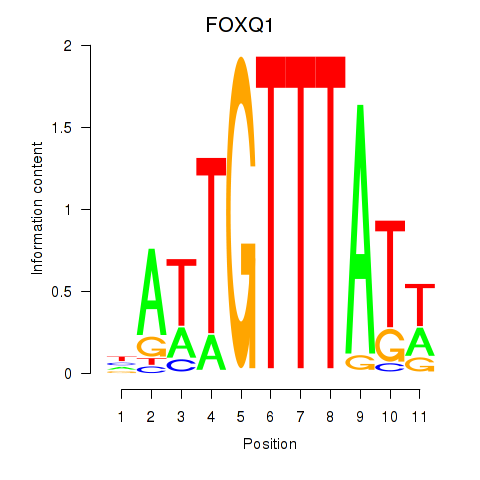
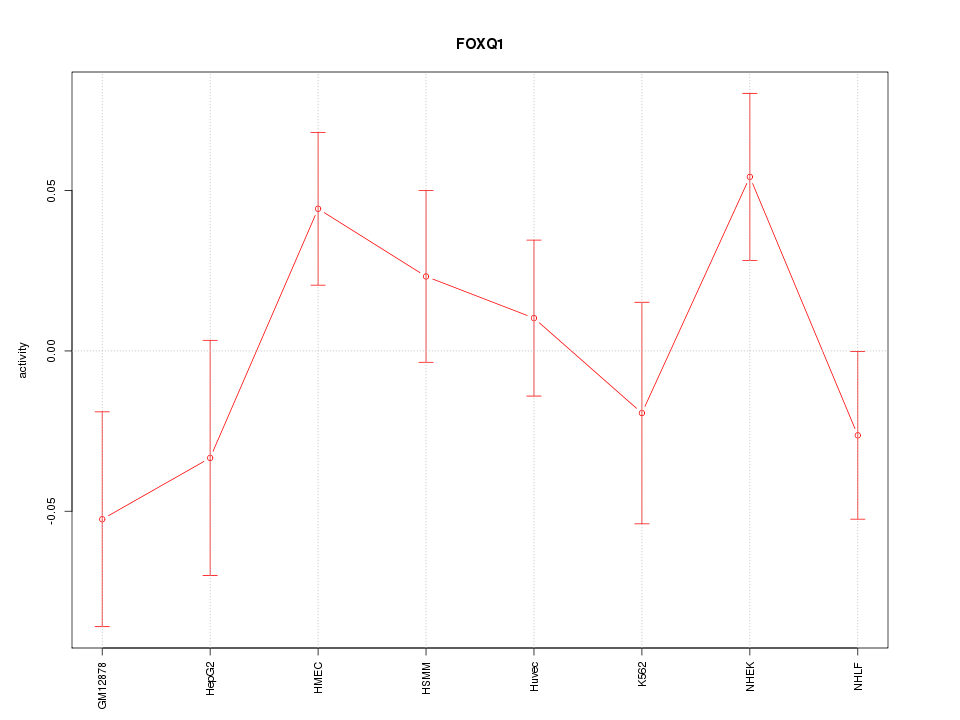
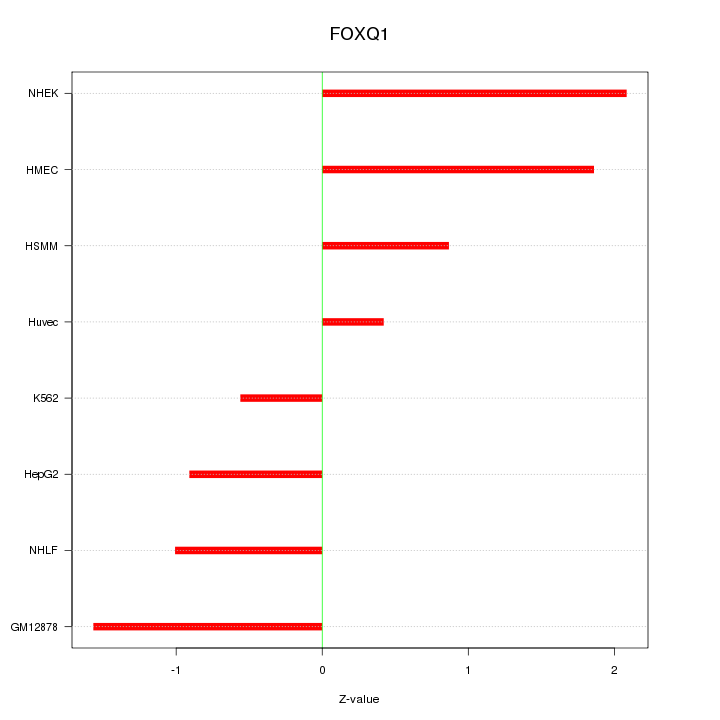
| 0.7 |
2.0 |
GO:0006015 |
5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.6 |
3.7 |
GO:0007499 |
ectoderm and mesoderm interaction(GO:0007499) |
| 0.4 |
1.3 |
GO:0060279 |
progesterone secretion(GO:0042701) regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.4 |
1.3 |
GO:0010837 |
regulation of keratinocyte proliferation(GO:0010837) |
| 0.3 |
0.9 |
GO:0042759 |
fatty acid elongation, saturated fatty acid(GO:0019367) long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.3 |
7.3 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 |
1.5 |
GO:0006880 |
intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 |
1.1 |
GO:0070050 |
neuron cellular homeostasis(GO:0070050) |
| 0.2 |
1.0 |
GO:0050703 |
interleukin-1 alpha secretion(GO:0050703) |
| 0.2 |
1.2 |
GO:0060242 |
contact inhibition(GO:0060242) |
| 0.2 |
0.4 |
GO:0045605 |
negative regulation of epidermal cell differentiation(GO:0045605) |
| 0.2 |
0.9 |
GO:0051902 |
negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.1 |
0.8 |
GO:0032472 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) Golgi calcium ion transport(GO:0032472) manganese ion homeostasis(GO:0055071) |
| 0.1 |
1.3 |
GO:0043497 |
regulation of protein heterodimerization activity(GO:0043497) |
| 0.1 |
0.6 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.1 |
0.4 |
GO:0001999 |
renal response to blood flow involved in circulatory renin-angiotensin regulation of systemic arterial blood pressure(GO:0001999) |
| 0.1 |
0.6 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.1 |
0.3 |
GO:0060028 |
convergent extension involved in axis elongation(GO:0060028) |
| 0.1 |
0.4 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 |
0.6 |
GO:2000111 |
positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.1 |
0.2 |
GO:0051088 |
PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 |
0.2 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.1 |
0.3 |
GO:0048669 |
collateral sprouting in absence of injury(GO:0048669) |
| 0.1 |
0.4 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.1 |
0.4 |
GO:0033033 |
negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.1 |
0.2 |
GO:0006726 |
eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.1 |
0.3 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.1 |
2.0 |
GO:0060512 |
prostate gland morphogenesis(GO:0060512) |
| 0.1 |
0.2 |
GO:0046621 |
negative regulation of organ growth(GO:0046621) |
| 0.1 |
1.8 |
GO:0006309 |
DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.1 |
1.6 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.0 |
0.6 |
GO:0006469 |
negative regulation of protein kinase activity(GO:0006469) |
| 0.0 |
0.8 |
GO:1902749 |
regulation of G2/M transition of mitotic cell cycle(GO:0010389) regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 |
0.3 |
GO:0008653 |
lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 |
0.2 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 |
0.1 |
GO:0045719 |
negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 |
0.3 |
GO:0032527 |
retrograde protein transport, ER to cytosol(GO:0030970) protein exit from endoplasmic reticulum(GO:0032527) |
| 0.0 |
1.2 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.0 |
0.1 |
GO:0050747 |
positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 |
0.5 |
GO:0006743 |
ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 |
0.9 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
1.0 |
GO:0042094 |
interleukin-2 biosynthetic process(GO:0042094) |
| 0.0 |
0.3 |
GO:0033119 |
negative regulation of RNA splicing(GO:0033119) |
| 0.0 |
0.2 |
GO:0032780 |
negative regulation of ATPase activity(GO:0032780) |
| 0.0 |
0.2 |
GO:0002430 |
complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 |
0.1 |
GO:0002541 |
activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 |
0.5 |
GO:0042403 |
thyroid hormone metabolic process(GO:0042403) |
| 0.0 |
2.1 |
GO:0009411 |
response to UV(GO:0009411) |
| 0.0 |
0.2 |
GO:0044245 |
polysaccharide digestion(GO:0044245) |
| 0.0 |
0.9 |
GO:0030520 |
intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 |
0.7 |
GO:0034199 |
activation of protein kinase A activity(GO:0034199) |
| 0.0 |
0.2 |
GO:0045008 |
depyrimidination(GO:0045008) |
| 0.0 |
0.6 |
GO:0051180 |
vitamin transport(GO:0051180) |
| 0.0 |
0.4 |
GO:0030968 |
endoplasmic reticulum unfolded protein response(GO:0030968) cellular response to unfolded protein(GO:0034620) |
| 0.0 |
0.4 |
GO:0046627 |
negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 |
0.4 |
GO:0007172 |
signal complex assembly(GO:0007172) |
| 0.0 |
0.2 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.2 |
GO:0007171 |
activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 |
0.1 |
GO:0030007 |
cellular potassium ion homeostasis(GO:0030007) negative regulation of cell volume(GO:0045794) |
| 0.0 |
0.1 |
GO:0071400 |
cellular response to oleic acid(GO:0071400) |
| 0.0 |
1.5 |
GO:0007163 |
establishment or maintenance of cell polarity(GO:0007163) |
| 0.0 |
0.1 |
GO:0001969 |
activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) positive regulation of activation of membrane attack complex(GO:0001970) positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 |
0.3 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.0 |
0.1 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.1 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 |
0.2 |
GO:0007059 |
chromosome segregation(GO:0007059) |
| 0.0 |
0.8 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |
| 0.0 |
0.0 |
GO:1904396 |
regulation of synaptic growth at neuromuscular junction(GO:0008582) regulation of neuromuscular junction development(GO:1904396) |
| 0.0 |
0.5 |
GO:0007043 |
cell-cell junction assembly(GO:0007043) |