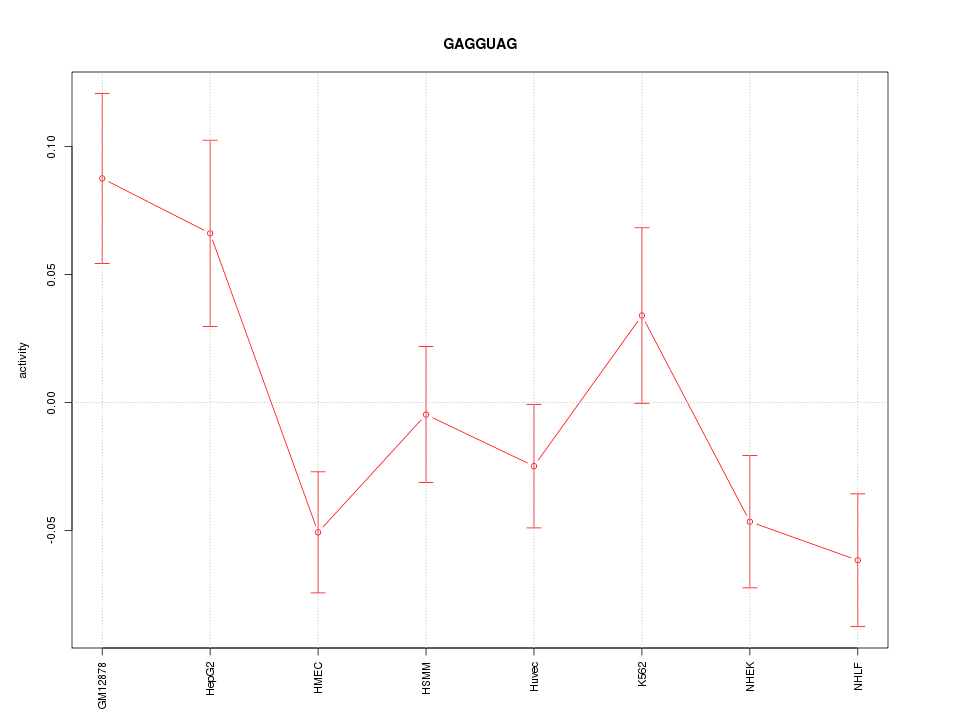
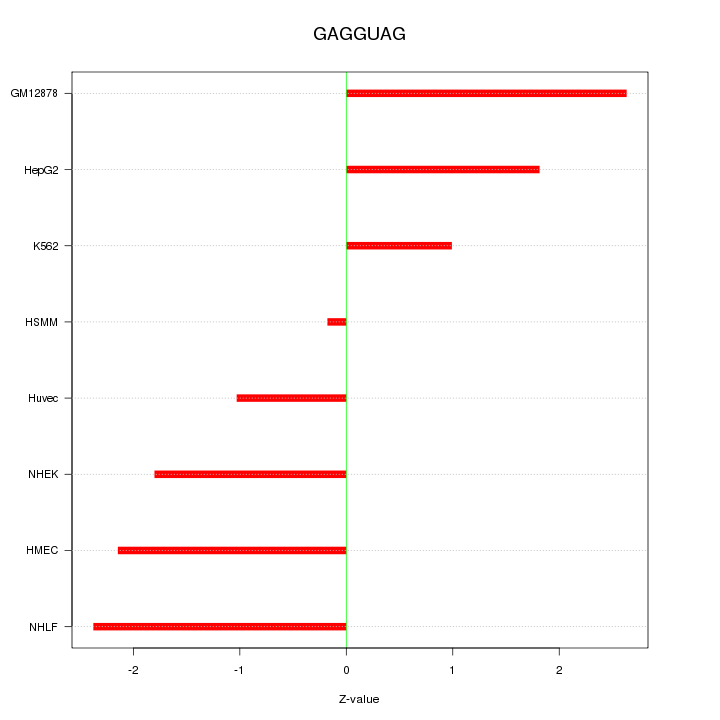
Motif ID: GAGGUAG
Z-value: 1.796

Mature miRNA associated with seed GAGGUAG:
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0060318 | definitive erythrocyte differentiation(GO:0060318) |
| 0.7 | 2.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.5 | 1.5 | GO:0032328 | alanine transport(GO:0032328) |
| 0.5 | 1.5 | GO:0060267 | transformation of host cell by virus(GO:0019087) positive regulation of respiratory burst(GO:0060267) |
| 0.5 | 2.7 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.4 | 1.1 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.3 | 1.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.3 | 1.3 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.3 | 0.8 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.3 | 0.8 | GO:0061298 | progesterone secretion(GO:0042701) retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.2 | 0.9 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.2 | 0.7 | GO:0060547 | negative regulation of necrotic cell death(GO:0060547) |
| 0.2 | 0.8 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.2 | 0.6 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.2 | 1.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.2 | 1.1 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.2 | 0.8 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 0.5 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 0.2 | 0.5 | GO:0042097 | lymphotoxin A production(GO:0032641) interleukin-4 biosynthetic process(GO:0042097) lymphotoxin A biosynthetic process(GO:0042109) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.2 | 2.0 | GO:0007512 | adult heart development(GO:0007512) |
| 0.2 | 2.5 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.2 | 1.7 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 0.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.3 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.1 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.5 | GO:0071025 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.1 | 1.7 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 0.4 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.1 | 0.4 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.1 | 0.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.6 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.7 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.1 | 0.7 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.3 | GO:0046520 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.5 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 0.1 | GO:0071635 | negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.1 | 0.5 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 | 0.5 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 0.4 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 0.3 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.1 | 0.6 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.8 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.1 | 0.3 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 0.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.8 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 0.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.7 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.1 | 0.5 | GO:0060676 | ureteric bud formation(GO:0060676) mesonephric tubule formation(GO:0072172) |
| 0.1 | 0.5 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 0.3 | GO:0046778 | modification by virus of host mRNA processing(GO:0046778) |
| 0.1 | 0.7 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.2 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 0.2 | GO:0003207 | cardiac chamber formation(GO:0003207) cardiac ventricle formation(GO:0003211) cardiac left ventricle formation(GO:0003218) |
| 0.1 | 0.4 | GO:0050685 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.1 | 0.7 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.1 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) DNA dealkylation(GO:0035510) |
| 0.1 | 0.6 | GO:0046051 | GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.1 | 0.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 2.0 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.1 | 0.3 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.1 | 0.4 | GO:0003148 | outflow tract septum morphogenesis(GO:0003148) |
| 0.1 | 1.1 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.1 | 0.3 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.1 | 0.3 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) |
| 0.1 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.8 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.5 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.1 | 0.6 | GO:0051567 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.0 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.2 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.4 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.1 | GO:0070544 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.3 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.2 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.1 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.4 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.0 | GO:0045069 | regulation of viral genome replication(GO:0045069) |
| 0.0 | 0.4 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.5 | GO:0001573 | ganglioside metabolic process(GO:0001573) placenta blood vessel development(GO:0060674) |
| 0.0 | 0.6 | GO:1902603 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.1 | GO:0010834 | obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 | 0.7 | GO:0006386 | transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 | 0.6 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 1.1 | GO:0051925 | obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.0 | 0.5 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.1 | GO:0010586 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.2 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.7 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 | 1.5 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.7 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0010523 | negative regulation of calcium ion transport into cytosol(GO:0010523) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.3 | GO:0034644 | cellular response to UV(GO:0034644) |
| 0.0 | 0.1 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.0 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.5 | GO:0006023 | aminoglycan biosynthetic process(GO:0006023) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.2 | GO:0001660 | fever generation(GO:0001660) |
| 0.0 | 0.9 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.1 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.3 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.6 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 1.6 | GO:0070304 | positive regulation of stress-activated MAPK cascade(GO:0032874) positive regulation of stress-activated protein kinase signaling cascade(GO:0070304) |
| 0.0 | 0.1 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.9 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.3 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 1.5 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.2 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.8 | GO:0050868 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 0.3 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.4 | GO:0036503 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) ERAD pathway(GO:0036503) |
| 0.0 | 1.1 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.2 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.4 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.2 | GO:0040023 | establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.3 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 2.3 | GO:0050658 | nucleic acid transport(GO:0050657) RNA transport(GO:0050658) establishment of RNA localization(GO:0051236) |
| 0.0 | 0.2 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.4 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 1.0 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.2 | GO:0050812 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.1 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.2 | GO:0009755 | hormone-mediated signaling pathway(GO:0009755) |
| 0.0 | 0.1 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.3 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) |
| 0.0 | 0.2 | GO:0034453 | microtubule nucleation(GO:0007020) microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 0.0 | 1.7 | GO:0007204 | positive regulation of cytosolic calcium ion concentration(GO:0007204) |
| 0.0 | 0.4 | GO:0016579 | protein deubiquitination(GO:0016579) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.4 | 1.6 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.4 | 1.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.3 | 1.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.3 | 1.4 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.2 | 1.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 1.2 | GO:0001741 | XY body(GO:0001741) |
| 0.2 | 0.7 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 0.5 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.1 | 0.7 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 1.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.0 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 0.3 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.1 | 0.5 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 0.7 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.1 | 1.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.5 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.2 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 2.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.8 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0009279 | cell outer membrane(GO:0009279) |
| 0.0 | 2.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.0 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 1.1 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.6 | GO:0090568 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.3 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.5 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 1.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.0 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) PAS complex(GO:0070772) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) nuclear outer membrane-endoplasmic reticulum membrane network(GO:0042175) |
| 0.0 | 0.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.6 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.3 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 1.4 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.5 | 1.5 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.5 | 1.5 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.4 | 0.4 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.4 | 1.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.4 | 1.1 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.3 | 0.8 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.2 | 0.7 | GO:0015217 | purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 1.7 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.2 | 0.8 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 1.4 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.2 | 1.9 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.2 | 0.5 | GO:0032422 | translation repressor activity, nucleic acid binding(GO:0000900) purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 2.7 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.2 | 0.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) glycine:sodium symporter activity(GO:0015375) |
| 0.2 | 1.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.2 | 0.5 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.2 | 0.6 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.1 | 1.0 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.1 | 1.2 | GO:0004835 | tubulin-tyrosine ligase activity(GO:0004835) |
| 0.1 | 0.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.6 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.6 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.8 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.1 | 0.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 1.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.5 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.1 | 0.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.3 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.3 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 2.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.4 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.6 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.1 | 2.0 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 0.8 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.1 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.2 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.1 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.2 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 1.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 1.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.4 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 1.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0004741 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.1 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.1 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 2.0 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 1.1 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.3 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 1.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.2 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.0 | GO:0070700 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) BMP receptor binding(GO:0070700) |
| 0.0 | 3.6 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.9 | GO:0004221 | obsolete ubiquitin thiolesterase activity(GO:0004221) |
| 0.0 | 0.5 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 1.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.7 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 2.1 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.5 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 2.6 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.7 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.3 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 10.4 | GO:0003700 | transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.6 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.2 | SIG_IL4RECEPTOR_IN_B_LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.6 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.6 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.7 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.2 | ST_JAK_STAT_PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.1 | ST_IL_13_PATHWAY | Interleukin 13 (IL-13) Pathway |