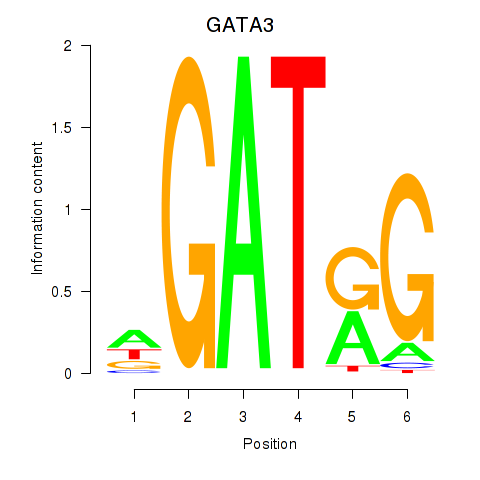
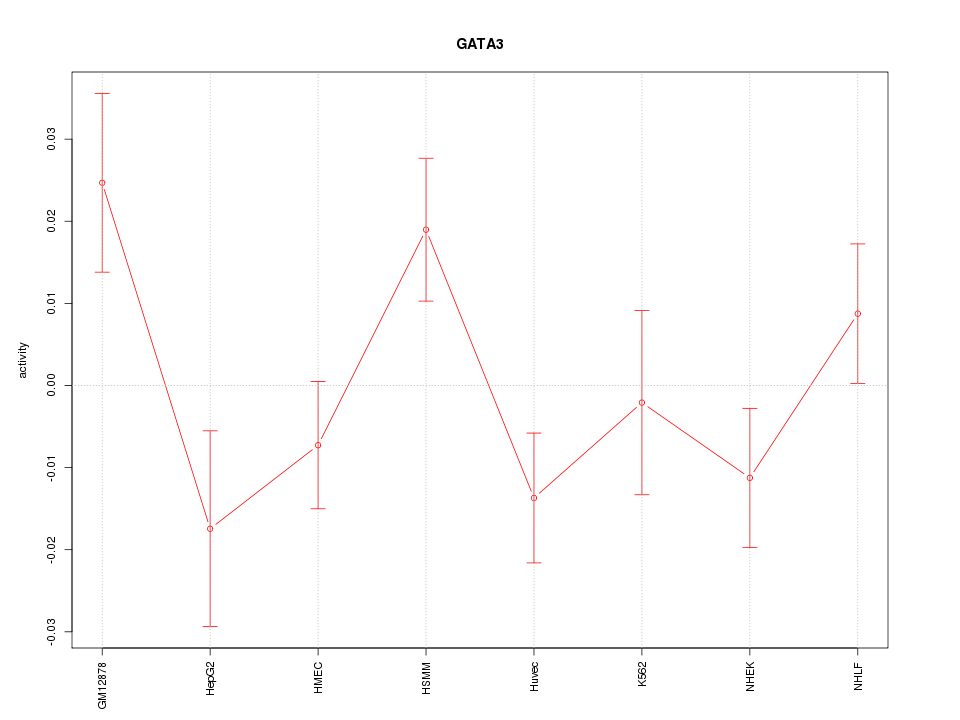
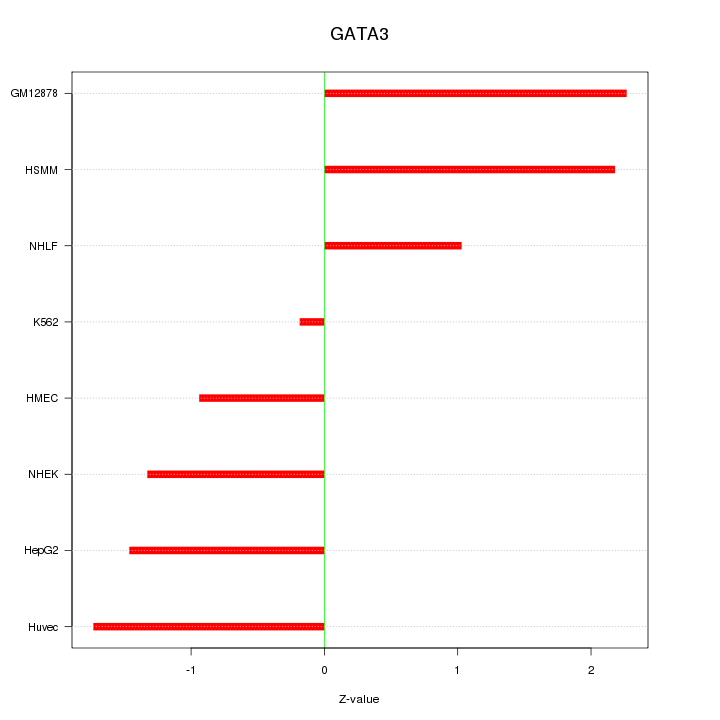
| 0.8 |
2.5 |
GO:0035408 |
histone H3-T6 phosphorylation(GO:0035408) |
| 0.6 |
0.6 |
GO:0051902 |
negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.5 |
0.5 |
GO:0006942 |
regulation of striated muscle contraction(GO:0006942) |
| 0.5 |
3.3 |
GO:0030007 |
cellular potassium ion homeostasis(GO:0030007) negative regulation of cell volume(GO:0045794) |
| 0.4 |
1.5 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 0.3 |
1.3 |
GO:2001258 |
negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) negative regulation of calcium ion transmembrane transporter activity(GO:1901020) negative regulation of cation channel activity(GO:2001258) |
| 0.3 |
3.3 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.3 |
0.5 |
GO:0007412 |
axon target recognition(GO:0007412) |
| 0.3 |
0.3 |
GO:0002637 |
regulation of immunoglobulin production(GO:0002637) positive regulation of immunoglobulin production(GO:0002639) |
| 0.3 |
1.5 |
GO:0030186 |
melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.2 |
1.4 |
GO:0002726 |
positive regulation of T cell cytokine production(GO:0002726) |
| 0.2 |
0.7 |
GO:0061030 |
epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.2 |
0.9 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.2 |
2.1 |
GO:0070474 |
uterine smooth muscle contraction(GO:0070471) regulation of uterine smooth muscle contraction(GO:0070472) positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.2 |
0.7 |
GO:0090047 |
obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.2 |
0.7 |
GO:0060028 |
convergent extension involved in axis elongation(GO:0060028) |
| 0.2 |
0.2 |
GO:0035751 |
vacuolar acidification(GO:0007035) lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.2 |
0.4 |
GO:0051097 |
negative regulation of helicase activity(GO:0051097) |
| 0.2 |
5.5 |
GO:0002504 |
antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.2 |
0.6 |
GO:0060340 |
pericardium morphogenesis(GO:0003344) diencephalon morphogenesis(GO:0048852) positive regulation of type I interferon-mediated signaling pathway(GO:0060340) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.2 |
0.6 |
GO:0045062 |
extrathymic T cell differentiation(GO:0033078) extrathymic T cell selection(GO:0045062) |
| 0.2 |
1.0 |
GO:0070050 |
neuron cellular homeostasis(GO:0070050) |
| 0.2 |
0.5 |
GO:0032196 |
transposition, DNA-mediated(GO:0006313) transposition(GO:0032196) |
| 0.2 |
0.5 |
GO:0001845 |
phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 0.2 |
1.1 |
GO:1901881 |
positive regulation of protein depolymerization(GO:1901881) |
| 0.2 |
0.5 |
GO:0006015 |
5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 |
0.5 |
GO:0033364 |
mast cell secretory granule organization(GO:0033364) |
| 0.2 |
0.7 |
GO:0006780 |
uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.2 |
0.5 |
GO:0046586 |
regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.2 |
2.1 |
GO:1901797 |
negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.2 |
0.9 |
GO:0043985 |
histone H4-R3 methylation(GO:0043985) |
| 0.2 |
0.3 |
GO:0040037 |
negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.2 |
1.1 |
GO:0034616 |
response to laminar fluid shear stress(GO:0034616) |
| 0.2 |
0.9 |
GO:2000124 |
cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 |
1.2 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.1 |
0.3 |
GO:0060338 |
regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.1 |
0.8 |
GO:0030240 |
skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 |
0.4 |
GO:0051255 |
spindle midzone assembly(GO:0051255) meiotic spindle midzone assembly(GO:0051257) |
| 0.1 |
4.0 |
GO:0050853 |
B cell receptor signaling pathway(GO:0050853) |
| 0.1 |
1.1 |
GO:0042693 |
muscle cell fate commitment(GO:0042693) |
| 0.1 |
0.4 |
GO:0071529 |
cementum mineralization(GO:0071529) |
| 0.1 |
0.3 |
GO:0006104 |
succinyl-CoA metabolic process(GO:0006104) |
| 0.1 |
0.3 |
GO:0032242 |
regulation of nucleoside transport(GO:0032242) |
| 0.1 |
0.4 |
GO:0097300 |
regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.1 |
0.4 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.1 |
0.4 |
GO:0032713 |
negative regulation of interleukin-4 production(GO:0032713) |
| 0.1 |
0.7 |
GO:0022614 |
membrane to membrane docking(GO:0022614) |
| 0.1 |
0.7 |
GO:0070168 |
negative regulation of biomineral tissue development(GO:0070168) |
| 0.1 |
0.4 |
GO:0002689 |
negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.1 |
0.2 |
GO:0071380 |
cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 |
0.5 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 |
1.1 |
GO:0006030 |
chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 |
0.7 |
GO:0042483 |
negative regulation of odontogenesis(GO:0042483) |
| 0.1 |
0.6 |
GO:0002335 |
mature B cell differentiation involved in immune response(GO:0002313) mature B cell differentiation(GO:0002335) |
| 0.1 |
0.7 |
GO:0033160 |
positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.1 |
0.3 |
GO:0019731 |
antimicrobial humoral response(GO:0019730) antibacterial humoral response(GO:0019731) |
| 0.1 |
0.7 |
GO:0014834 |
skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.1 |
0.3 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.1 |
0.7 |
GO:0030952 |
establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.1 |
0.4 |
GO:0043128 |
regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.1 |
0.5 |
GO:0043116 |
negative regulation of vascular permeability(GO:0043116) |
| 0.1 |
0.8 |
GO:0045078 |
positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 |
0.4 |
GO:0021978 |
telencephalon regionalization(GO:0021978) |
| 0.1 |
0.6 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.1 |
0.5 |
GO:0022604 |
regulation of cell morphogenesis(GO:0022604) |
| 0.1 |
0.5 |
GO:0046952 |
ketone body catabolic process(GO:0046952) |
| 0.1 |
0.2 |
GO:0002291 |
T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 |
0.3 |
GO:0001757 |
somite specification(GO:0001757) |
| 0.1 |
1.9 |
GO:0045730 |
respiratory burst(GO:0045730) |
| 0.1 |
7.0 |
GO:0030049 |
muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 |
0.4 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 |
0.4 |
GO:0000089 |
mitotic metaphase(GO:0000089) |
| 0.1 |
0.2 |
GO:0010716 |
regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 |
0.2 |
GO:0006154 |
adenosine catabolic process(GO:0006154) |
| 0.1 |
0.2 |
GO:0010966 |
regulation of phosphate transport(GO:0010966) |
| 0.1 |
0.5 |
GO:0006772 |
thiamine metabolic process(GO:0006772) |
| 0.1 |
0.9 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.1 |
0.7 |
GO:0071353 |
cellular response to interleukin-4(GO:0071353) |
| 0.1 |
0.3 |
GO:0016048 |
detection of temperature stimulus(GO:0016048) |
| 0.1 |
0.3 |
GO:0071848 |
regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) TNFSF11-mediated signaling pathway(GO:0071847) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.1 |
0.1 |
GO:0018209 |
peptidyl-serine modification(GO:0018209) |
| 0.1 |
0.2 |
GO:0046685 |
response to arsenic-containing substance(GO:0046685) |
| 0.1 |
0.5 |
GO:0001554 |
luteolysis(GO:0001554) |
| 0.1 |
0.1 |
GO:0008015 |
circulatory system process(GO:0003013) blood circulation(GO:0008015) |
| 0.1 |
0.2 |
GO:0032641 |
lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.1 |
0.2 |
GO:0030205 |
dermatan sulfate metabolic process(GO:0030205) dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 |
0.2 |
GO:0032914 |
positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 |
0.2 |
GO:0060024 |
rhythmic synaptic transmission(GO:0060024) |
| 0.1 |
0.2 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 |
0.2 |
GO:0060398 |
regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.1 |
0.2 |
GO:0007079 |
mitotic chromosome movement towards spindle pole(GO:0007079) chromosome movement towards spindle pole(GO:0051305) |
| 0.1 |
0.1 |
GO:0009441 |
glycolate metabolic process(GO:0009441) |
| 0.1 |
1.5 |
GO:0001516 |
prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 |
0.1 |
GO:0008543 |
fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.1 |
0.2 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 |
0.1 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.1 |
1.6 |
GO:0000303 |
response to superoxide(GO:0000303) |
| 0.1 |
0.4 |
GO:0001562 |
response to protozoan(GO:0001562) |
| 0.1 |
0.4 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.1 |
0.2 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) cytoplasmic translation(GO:0002181) cytoplasmic translational initiation(GO:0002183) |
| 0.1 |
1.5 |
GO:0030199 |
collagen fibril organization(GO:0030199) |
| 0.1 |
0.7 |
GO:0007204 |
positive regulation of cytosolic calcium ion concentration(GO:0007204) |
| 0.1 |
0.4 |
GO:0042769 |
DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 |
0.4 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.1 |
0.9 |
GO:0045885 |
obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.1 |
0.2 |
GO:0051901 |
positive regulation of mitochondrial depolarization(GO:0051901) positive regulation of membrane depolarization(GO:1904181) |
| 0.1 |
0.7 |
GO:0001782 |
B cell homeostasis(GO:0001782) |
| 0.1 |
0.4 |
GO:0044413 |
evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 |
0.2 |
GO:0060295 |
regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) cilium movement involved in cell motility(GO:0060294) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 |
0.2 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.1 |
0.1 |
GO:0046967 |
cytosol to ER transport(GO:0046967) |
| 0.1 |
0.5 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 |
0.2 |
GO:0085029 |
extracellular matrix assembly(GO:0085029) |
| 0.1 |
0.4 |
GO:0006880 |
intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 |
0.1 |
GO:0006651 |
diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 |
0.2 |
GO:0034770 |
histone H4-K20 methylation(GO:0034770) |
| 0.1 |
0.2 |
GO:0045988 |
negative regulation of striated muscle contraction(GO:0045988) |
| 0.1 |
0.2 |
GO:0002478 |
antigen processing and presentation of exogenous peptide antigen(GO:0002478) |
| 0.1 |
0.2 |
GO:0010507 |
negative regulation of autophagy(GO:0010507) |
| 0.1 |
0.5 |
GO:0032814 |
regulation of natural killer cell activation(GO:0032814) |
| 0.1 |
0.2 |
GO:0051823 |
hindbrain radial glia guided cell migration(GO:0021932) regulation of synapse structural plasticity(GO:0051823) |
| 0.1 |
2.6 |
GO:0050690 |
regulation of defense response to virus by virus(GO:0050690) |
| 0.1 |
0.1 |
GO:0003084 |
positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.1 |
0.3 |
GO:0050435 |
beta-amyloid metabolic process(GO:0050435) |
| 0.1 |
0.1 |
GO:0031033 |
positive regulation of lamellipodium assembly(GO:0010592) myosin filament organization(GO:0031033) positive regulation of lamellipodium organization(GO:1902745) |
| 0.1 |
0.9 |
GO:0050930 |
induction of positive chemotaxis(GO:0050930) |
| 0.1 |
0.4 |
GO:0060155 |
platelet dense granule organization(GO:0060155) |
| 0.1 |
0.4 |
GO:0051895 |
negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 |
0.1 |
GO:0050862 |
positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 |
0.2 |
GO:0051924 |
regulation of calcium ion transport(GO:0051924) |
| 0.1 |
0.2 |
GO:0006782 |
protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.1 |
0.5 |
GO:0006975 |
DNA damage induced protein phosphorylation(GO:0006975) |
| 0.1 |
0.2 |
GO:0006435 |
threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 |
0.2 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.1 |
0.2 |
GO:0045900 |
negative regulation of translational elongation(GO:0045900) |
| 0.1 |
0.1 |
GO:0002040 |
sprouting angiogenesis(GO:0002040) |
| 0.1 |
1.2 |
GO:0008347 |
glial cell migration(GO:0008347) |
| 0.1 |
0.1 |
GO:0070922 |
small RNA loading onto RISC(GO:0070922) |
| 0.1 |
0.1 |
GO:0033262 |
regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.1 |
0.2 |
GO:0046340 |
diacylglycerol catabolic process(GO:0046340) |
| 0.1 |
0.3 |
GO:0006552 |
leucine catabolic process(GO:0006552) |
| 0.1 |
0.2 |
GO:0034086 |
maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.1 |
0.1 |
GO:0046813 |
receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.1 |
0.4 |
GO:0051383 |
kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.1 |
0.2 |
GO:0006669 |
sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 |
0.3 |
GO:0071480 |
cellular response to gamma radiation(GO:0071480) |
| 0.0 |
4.1 |
GO:0009408 |
response to heat(GO:0009408) |
| 0.0 |
0.3 |
GO:0046887 |
positive regulation of hormone secretion(GO:0046887) |
| 0.0 |
0.3 |
GO:0048486 |
parasympathetic nervous system development(GO:0048486) |
| 0.0 |
0.3 |
GO:0045084 |
positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 |
0.3 |
GO:0015742 |
alpha-ketoglutarate transport(GO:0015742) |
| 0.0 |
0.1 |
GO:0007285 |
primary spermatocyte growth(GO:0007285) |
| 0.0 |
0.4 |
GO:0070189 |
kynurenine metabolic process(GO:0070189) |
| 0.0 |
0.4 |
GO:0097061 |
dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 |
0.0 |
GO:0060746 |
maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 |
0.1 |
GO:1904396 |
regulation of synaptic growth at neuromuscular junction(GO:0008582) regulation of neuromuscular junction development(GO:1904396) |
| 0.0 |
0.2 |
GO:0015802 |
basic amino acid transport(GO:0015802) |
| 0.0 |
0.3 |
GO:0010759 |
regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) |
| 0.0 |
0.2 |
GO:0001831 |
trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 |
0.4 |
GO:0007185 |
transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 |
0.6 |
GO:0070206 |
protein trimerization(GO:0070206) |
| 0.0 |
0.1 |
GO:0031282 |
regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 |
0.1 |
GO:0030539 |
male genitalia development(GO:0030539) |
| 0.0 |
0.1 |
GO:0046950 |
cellular ketone body metabolic process(GO:0046950) |
| 0.0 |
0.2 |
GO:0032811 |
negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of epinephrine secretion(GO:0032811) negative regulation of dopamine secretion(GO:0033602) |
| 0.0 |
0.2 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 |
0.0 |
GO:0006353 |
DNA-templated transcription, termination(GO:0006353) |
| 0.0 |
0.2 |
GO:0016064 |
immunoglobulin mediated immune response(GO:0016064) |
| 0.0 |
0.1 |
GO:0071460 |
cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 |
0.2 |
GO:0060052 |
neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 |
0.1 |
GO:0050758 |
thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 |
0.1 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
0.5 |
GO:0030101 |
natural killer cell activation(GO:0030101) |
| 0.0 |
0.2 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 |
0.2 |
GO:0043923 |
positive regulation by host of viral transcription(GO:0043923) |
| 0.0 |
0.2 |
GO:0042535 |
positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 |
0.1 |
GO:0050685 |
positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 |
0.3 |
GO:0032647 |
interferon-alpha production(GO:0032607) regulation of interferon-alpha production(GO:0032647) positive regulation of interferon-alpha production(GO:0032727) |
| 0.0 |
0.8 |
GO:0033209 |
tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 |
0.2 |
GO:0031635 |
adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) opioid receptor signaling pathway(GO:0038003) |
| 0.0 |
0.1 |
GO:0033158 |
regulation of protein import into nucleus, translocation(GO:0033158) |
| 0.0 |
0.3 |
GO:0033261 |
obsolete regulation of S phase(GO:0033261) |
| 0.0 |
0.2 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.0 |
0.6 |
GO:0006941 |
striated muscle contraction(GO:0006941) |
| 0.0 |
0.2 |
GO:0070537 |
histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 |
0.1 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.0 |
0.2 |
GO:0006658 |
phosphatidylserine metabolic process(GO:0006658) |
| 0.0 |
0.0 |
GO:0031223 |
mechanosensory behavior(GO:0007638) response to auditory stimulus(GO:0010996) auditory behavior(GO:0031223) |
| 0.0 |
0.3 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.1 |
GO:0042518 |
negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 |
0.2 |
GO:0007182 |
common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 |
0.3 |
GO:0006188 |
IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.0 |
0.1 |
GO:0045671 |
negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 |
1.0 |
GO:0060334 |
regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 |
0.3 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.0 |
0.2 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 |
0.1 |
GO:0048241 |
epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) |
| 0.0 |
0.0 |
GO:0033043 |
regulation of organelle organization(GO:0033043) |
| 0.0 |
0.3 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.2 |
GO:0014819 |
regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 |
0.8 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.1 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.0 |
0.1 |
GO:0015917 |
aminophospholipid transport(GO:0015917) |
| 0.0 |
0.5 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 |
0.2 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.0 |
0.1 |
GO:0006428 |
isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 |
0.6 |
GO:0045069 |
regulation of viral genome replication(GO:0045069) |
| 0.0 |
0.1 |
GO:0001544 |
initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 |
0.7 |
GO:0034622 |
cellular macromolecular complex assembly(GO:0034622) |
| 0.0 |
0.1 |
GO:0006273 |
lagging strand elongation(GO:0006273) |
| 0.0 |
0.1 |
GO:0042532 |
negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 |
0.4 |
GO:0046697 |
decidualization(GO:0046697) |
| 0.0 |
0.1 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 |
0.1 |
GO:0006382 |
adenosine to inosine editing(GO:0006382) |
| 0.0 |
0.3 |
GO:0051249 |
regulation of lymphocyte activation(GO:0051249) |
| 0.0 |
0.1 |
GO:0006216 |
cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 |
0.7 |
GO:0030225 |
macrophage differentiation(GO:0030225) |
| 0.0 |
0.1 |
GO:0048745 |
smooth muscle tissue development(GO:0048745) |
| 0.0 |
0.1 |
GO:0060292 |
long term synaptic depression(GO:0060292) |
| 0.0 |
0.1 |
GO:0032506 |
cytokinetic process(GO:0032506) |
| 0.0 |
0.2 |
GO:0033523 |
histone H2B ubiquitination(GO:0033523) |
| 0.0 |
0.2 |
GO:0032329 |
serine transport(GO:0032329) |
| 0.0 |
0.2 |
GO:0015793 |
glycerol transport(GO:0015793) |
| 0.0 |
0.0 |
GO:0003323 |
type B pancreatic cell development(GO:0003323) |
| 0.0 |
0.1 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 |
0.1 |
GO:0048861 |
leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 |
0.7 |
GO:0009083 |
branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
0.3 |
GO:0035590 |
purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 |
0.1 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.0 |
0.1 |
GO:1901998 |
tetracycline transport(GO:0015904) antibiotic transport(GO:0042891) toxin transport(GO:1901998) |
| 0.0 |
0.4 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.1 |
GO:1902803 |
regulation of synaptic vesicle transport(GO:1902803) regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 |
0.2 |
GO:0001880 |
Mullerian duct regression(GO:0001880) |
| 0.0 |
4.0 |
GO:0065004 |
protein-DNA complex assembly(GO:0065004) |
| 0.0 |
0.1 |
GO:0031290 |
retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 |
0.5 |
GO:0031532 |
actin cytoskeleton reorganization(GO:0031532) |
| 0.0 |
0.2 |
GO:0015755 |
fructose transport(GO:0015755) |
| 0.0 |
0.1 |
GO:0048711 |
positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 |
0.1 |
GO:0001941 |
postsynaptic membrane organization(GO:0001941) |
| 0.0 |
0.7 |
GO:0018208 |
peptidyl-proline modification(GO:0018208) |
| 0.0 |
0.9 |
GO:0002286 |
T cell activation involved in immune response(GO:0002286) |
| 0.0 |
0.1 |
GO:0060363 |
cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 |
0.2 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 |
0.1 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 |
0.4 |
GO:0048536 |
spleen development(GO:0048536) |
| 0.0 |
0.3 |
GO:0051482 |
positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 |
0.0 |
GO:0031953 |
negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 |
0.2 |
GO:0008608 |
attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 |
0.1 |
GO:0045199 |
maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 |
0.1 |
GO:0014044 |
Schwann cell development(GO:0014044) myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 |
0.0 |
GO:0032740 |
positive regulation of interleukin-17 production(GO:0032740) |
| 0.0 |
0.1 |
GO:0072393 |
microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 |
0.1 |
GO:0060744 |
thelarche(GO:0042695) development of secondary female sexual characteristics(GO:0046543) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 |
0.4 |
GO:0051496 |
positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 |
0.1 |
GO:0071436 |
sodium ion export(GO:0071436) |
| 0.0 |
0.1 |
GO:0061101 |
neuroendocrine cell differentiation(GO:0061101) |
| 0.0 |
0.1 |
GO:0009136 |
ADP biosynthetic process(GO:0006172) nucleoside diphosphate biosynthetic process(GO:0009133) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 |
0.1 |
GO:0010761 |
fibroblast migration(GO:0010761) |
| 0.0 |
0.1 |
GO:0071428 |
rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 |
0.1 |
GO:0006122 |
mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 |
0.0 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 |
0.2 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.0 |
0.1 |
GO:0042262 |
DNA protection(GO:0042262) |
| 0.0 |
0.1 |
GO:0006069 |
ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 |
0.5 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.1 |
GO:0048630 |
skeletal muscle tissue growth(GO:0048630) |
| 0.0 |
0.2 |
GO:0045540 |
regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 |
0.1 |
GO:0010535 |
regulation of activation of JAK2 kinase activity(GO:0010534) positive regulation of activation of JAK2 kinase activity(GO:0010535) |
| 0.0 |
0.1 |
GO:0018916 |
nitrobenzene metabolic process(GO:0018916) |
| 0.0 |
0.3 |
GO:0043403 |
skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 |
0.1 |
GO:0072136 |
kidney mesenchymal cell proliferation(GO:0072135) metanephric mesenchymal cell proliferation involved in metanephros development(GO:0072136) |
| 0.0 |
0.3 |
GO:0042531 |
positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 |
0.2 |
GO:0000090 |
mitotic anaphase(GO:0000090) |
| 0.0 |
0.1 |
GO:0090050 |
regulation of cell migration involved in sprouting angiogenesis(GO:0090049) positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) regulation of sprouting angiogenesis(GO:1903670) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 |
0.3 |
GO:0046051 |
GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) pyrimidine ribonucleoside biosynthetic process(GO:0046132) |
| 0.0 |
0.1 |
GO:0033632 |
regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 |
0.2 |
GO:0042059 |
negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 |
0.2 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.0 |
0.9 |
GO:0042771 |
intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 |
0.3 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 |
0.2 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.0 |
0.1 |
GO:0015824 |
proline transport(GO:0015824) |
| 0.0 |
0.3 |
GO:0071479 |
cellular response to ionizing radiation(GO:0071479) |
| 0.0 |
0.1 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 |
0.1 |
GO:0051563 |
smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 |
0.2 |
GO:0046638 |
regulation of alpha-beta T cell differentiation(GO:0046637) positive regulation of alpha-beta T cell differentiation(GO:0046638) |
| 0.0 |
0.0 |
GO:0042415 |
norepinephrine metabolic process(GO:0042415) |
| 0.0 |
0.0 |
GO:0046133 |
pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.0 |
1.5 |
GO:0006338 |
chromatin remodeling(GO:0006338) |
| 0.0 |
1.2 |
GO:0060333 |
interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 |
0.0 |
GO:0009148 |
pyrimidine nucleoside triphosphate biosynthetic process(GO:0009148) |
| 0.0 |
0.6 |
GO:0015949 |
nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 |
0.4 |
GO:0035195 |
gene silencing by miRNA(GO:0035195) |
| 0.0 |
0.0 |
GO:1903320 |
regulation of protein modification by small protein conjugation or removal(GO:1903320) |
| 0.0 |
0.4 |
GO:0008625 |
extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 |
0.1 |
GO:0042219 |
cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 |
0.2 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.0 |
0.1 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.0 |
0.2 |
GO:0007063 |
regulation of sister chromatid cohesion(GO:0007063) |
| 0.0 |
0.0 |
GO:1903960 |
negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 |
0.5 |
GO:1902749 |
regulation of G2/M transition of mitotic cell cycle(GO:0010389) regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 |
0.1 |
GO:0071168 |
protein localization to chromatin(GO:0071168) |
| 0.0 |
2.2 |
GO:0042113 |
B cell activation(GO:0042113) |
| 0.0 |
0.1 |
GO:0046653 |
tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 |
0.2 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 |
0.4 |
GO:0006044 |
N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 |
0.1 |
GO:0010898 |
positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 |
0.1 |
GO:0070493 |
thrombin receptor signaling pathway(GO:0070493) |
| 0.0 |
0.2 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) |
| 0.0 |
0.0 |
GO:0051310 |
mitotic metaphase plate congression(GO:0007080) metaphase plate congression(GO:0051310) |
| 0.0 |
0.5 |
GO:0043101 |
purine-containing compound salvage(GO:0043101) |
| 0.0 |
0.1 |
GO:0090231 |
regulation of spindle checkpoint(GO:0090231) |
| 0.0 |
0.1 |
GO:0071636 |
transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) positive regulation of transforming growth factor beta production(GO:0071636) |
| 0.0 |
0.1 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 |
0.1 |
GO:0001906 |
cell killing(GO:0001906) |
| 0.0 |
0.1 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.0 |
3.4 |
GO:0043123 |
positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 |
0.1 |
GO:0050884 |
neuromuscular process controlling posture(GO:0050884) |
| 0.0 |
0.4 |
GO:0019079 |
viral genome replication(GO:0019079) |
| 0.0 |
0.1 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.0 |
4.6 |
GO:0007517 |
muscle organ development(GO:0007517) |
| 0.0 |
0.1 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.0 |
GO:0010002 |
cardioblast differentiation(GO:0010002) |
| 0.0 |
0.0 |
GO:0060972 |
left/right pattern formation(GO:0060972) |
| 0.0 |
0.1 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.1 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 |
0.0 |
GO:0007178 |
transmembrane receptor protein serine/threonine kinase signaling pathway(GO:0007178) |
| 0.0 |
0.2 |
GO:0031529 |
ruffle organization(GO:0031529) |
| 0.0 |
0.1 |
GO:0016447 |
somatic recombination of immunoglobulin gene segments(GO:0016447) |
| 0.0 |
0.0 |
GO:0000183 |
chromatin silencing at rDNA(GO:0000183) |
| 0.0 |
0.0 |
GO:0009103 |
lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 |
0.3 |
GO:0030851 |
granulocyte differentiation(GO:0030851) |
| 0.0 |
0.1 |
GO:0009106 |
lipoate metabolic process(GO:0009106) lipoate biosynthetic process(GO:0009107) |
| 0.0 |
0.1 |
GO:0009109 |
coenzyme catabolic process(GO:0009109) |
| 0.0 |
0.1 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.0 |
0.0 |
GO:0051968 |
positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 |
0.2 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.0 |
0.0 |
GO:0048845 |
venous blood vessel morphogenesis(GO:0048845) |
| 0.0 |
0.1 |
GO:0035608 |
protein deglutamylation(GO:0035608) |
| 0.0 |
0.3 |
GO:0051131 |
chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 |
0.0 |
GO:0030185 |
nitric oxide transport(GO:0030185) |
| 0.0 |
0.1 |
GO:0006611 |
protein export from nucleus(GO:0006611) |
| 0.0 |
0.0 |
GO:0051365 |
cellular response to potassium ion starvation(GO:0051365) |
| 0.0 |
0.2 |
GO:0007162 |
negative regulation of cell adhesion(GO:0007162) |
| 0.0 |
0.1 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.0 |
0.3 |
GO:0030048 |
actin filament-based movement(GO:0030048) |
| 0.0 |
0.0 |
GO:0045648 |
positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 |
0.2 |
GO:0010874 |
regulation of cholesterol efflux(GO:0010874) |
| 0.0 |
0.1 |
GO:0014037 |
Schwann cell differentiation(GO:0014037) |
| 0.0 |
0.1 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.0 |
0.5 |
GO:0031032 |
actomyosin structure organization(GO:0031032) |
| 0.0 |
0.2 |
GO:0006734 |
NADH metabolic process(GO:0006734) |
| 0.0 |
0.1 |
GO:0030814 |
regulation of cAMP metabolic process(GO:0030814) |
| 0.0 |
0.1 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 |
0.1 |
GO:0007189 |
adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 |
0.1 |
GO:0032781 |
positive regulation of ATPase activity(GO:0032781) |
| 0.0 |
0.0 |
GO:0051154 |
negative regulation of myotube differentiation(GO:0010832) negative regulation of striated muscle cell differentiation(GO:0051154) |
| 0.0 |
0.2 |
GO:0034260 |
negative regulation of GTPase activity(GO:0034260) |
| 0.0 |
0.1 |
GO:0031167 |
rRNA methylation(GO:0031167) |
| 0.0 |
1.3 |
GO:0006977 |
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
| 0.0 |
0.1 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.0 |
0.2 |
GO:0000060 |
protein import into nucleus, translocation(GO:0000060) |
| 0.0 |
0.2 |
GO:0008634 |
obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.0 |
0.2 |
GO:0021513 |
spinal cord dorsal/ventral patterning(GO:0021513) |
| 0.0 |
0.0 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.0 |
GO:0042986 |
positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 |
0.1 |
GO:0010799 |
regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 |
0.1 |
GO:0007029 |
endoplasmic reticulum organization(GO:0007029) |
| 0.0 |
0.1 |
GO:0032527 |
retrograde protein transport, ER to cytosol(GO:0030970) protein exit from endoplasmic reticulum(GO:0032527) |
| 0.0 |
0.9 |
GO:0006892 |
post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 |
0.1 |
GO:0031664 |
regulation of lipopolysaccharide-mediated signaling pathway(GO:0031664) negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 |
0.1 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.0 |
0.1 |
GO:1902582 |
single-organism intracellular transport(GO:1902582) |
| 0.0 |
0.0 |
GO:0051546 |
keratinocyte migration(GO:0051546) |
| 0.0 |
0.1 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 |
0.1 |
GO:0015705 |
iodide transport(GO:0015705) |
| 0.0 |
0.0 |
GO:0071357 |
response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 |
0.1 |
GO:0009750 |
response to fructose(GO:0009750) |
| 0.0 |
0.1 |
GO:0007341 |
penetration of zona pellucida(GO:0007341) |
| 0.0 |
0.1 |
GO:0099515 |
nuclear migration along microfilament(GO:0031022) actin filament-based transport(GO:0099515) |
| 0.0 |
0.1 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.0 |
0.4 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.1 |
GO:0042977 |
activation of JAK2 kinase activity(GO:0042977) |
| 0.0 |
0.1 |
GO:0045007 |
depurination(GO:0045007) |
| 0.0 |
0.7 |
GO:0006333 |
chromatin assembly or disassembly(GO:0006333) |
| 0.0 |
0.0 |
GO:0031214 |
biomineral tissue development(GO:0031214) |
| 0.0 |
0.0 |
GO:0051971 |
positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.0 |
0.1 |
GO:0005984 |
disaccharide metabolic process(GO:0005984) |
| 0.0 |
0.1 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
0.1 |
GO:0031274 |
pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 |
0.1 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.3 |
GO:0006309 |
DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.0 |
0.1 |
GO:0032099 |
negative regulation of appetite(GO:0032099) |
| 0.0 |
0.5 |
GO:0032480 |
negative regulation of type I interferon production(GO:0032480) |
| 0.0 |
0.0 |
GO:0045744 |
negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 |
0.3 |
GO:0048255 |
RNA stabilization(GO:0043489) mRNA stabilization(GO:0048255) |
| 0.0 |
0.1 |
GO:0007346 |
regulation of mitotic cell cycle(GO:0007346) |
| 0.0 |
0.1 |
GO:0035964 |
COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 |
0.0 |
GO:0046292 |
formaldehyde metabolic process(GO:0046292) |
| 0.0 |
0.0 |
GO:0045838 |
positive regulation of membrane potential(GO:0045838) |
| 0.0 |
0.1 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 |
0.4 |
GO:0033572 |
ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 |
0.4 |
GO:0031397 |
negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 |
0.1 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.0 |
0.0 |
GO:0032464 |
positive regulation of protein oligomerization(GO:0032461) positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 |
0.1 |
GO:0010658 |
striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 |
0.2 |
GO:0042267 |
natural killer cell mediated immunity(GO:0002228) natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 |
0.1 |
GO:0010737 |
protein kinase A signaling(GO:0010737) regulation of protein kinase A signaling(GO:0010738) |
| 0.0 |
0.1 |
GO:1902603 |
carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 |
0.1 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.0 |
0.1 |
GO:0010157 |
response to chlorate(GO:0010157) |
| 0.0 |
0.3 |
GO:0045776 |
negative regulation of blood pressure(GO:0045776) |
| 0.0 |
0.0 |
GO:0009162 |
nucleoside monophosphate catabolic process(GO:0009125) deoxyribonucleoside monophosphate metabolic process(GO:0009162) pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) dUMP metabolic process(GO:0046078) |
| 0.0 |
0.1 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.0 |
0.2 |
GO:0008214 |
protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 |
0.8 |
GO:0051291 |
protein heterooligomerization(GO:0051291) |
| 0.0 |
0.0 |
GO:0019853 |
L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 |
0.2 |
GO:0006298 |
mismatch repair(GO:0006298) |
| 0.0 |
0.1 |
GO:1901070 |
GMP biosynthetic process(GO:0006177) guanosine-containing compound biosynthetic process(GO:1901070) |
| 0.0 |
0.0 |
GO:0032836 |
glomerular basement membrane development(GO:0032836) |
| 0.0 |
0.1 |
GO:0006108 |
malate metabolic process(GO:0006108) |
| 0.0 |
0.2 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.1 |
GO:0016998 |
cell wall macromolecule catabolic process(GO:0016998) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.0 |
0.0 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.0 |
0.1 |
GO:0060122 |
auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 |
0.1 |
GO:0000959 |
mitochondrial RNA metabolic process(GO:0000959) transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.0 |
GO:0007051 |
spindle organization(GO:0007051) |
| 0.0 |
0.3 |
GO:0030866 |
cortical cytoskeleton organization(GO:0030865) cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 |
0.0 |
GO:0042339 |
keratan sulfate metabolic process(GO:0042339) |
| 0.0 |
0.0 |
GO:0001552 |
ovarian follicle atresia(GO:0001552) |
| 0.0 |
0.1 |
GO:0006111 |
regulation of gluconeogenesis(GO:0006111) |
| 0.0 |
0.0 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 |
0.1 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.0 |
0.1 |
GO:0042036 |
negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 |
0.1 |
GO:0007612 |
learning(GO:0007612) |
| 0.0 |
0.0 |
GO:0031118 |
rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 |
0.0 |
GO:0051771 |
negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 |
0.1 |
GO:0006144 |
purine nucleobase metabolic process(GO:0006144) |
| 0.0 |
0.0 |
GO:0034587 |
RNA 5'-end processing(GO:0000966) piRNA metabolic process(GO:0034587) |
| 0.0 |
0.1 |
GO:0006529 |
asparagine biosynthetic process(GO:0006529) |
| 0.0 |
0.2 |
GO:0046655 |
folic acid metabolic process(GO:0046655) |
| 0.0 |
0.1 |
GO:0030321 |
transepithelial chloride transport(GO:0030321) |
| 0.0 |
0.5 |
GO:0006888 |
ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 |
0.6 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.0 |
0.0 |
GO:0006655 |
phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 |
0.1 |
GO:0051930 |
regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 |
0.0 |
GO:0060152 |
peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 |
0.0 |
GO:0072677 |
eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 |
0.1 |
GO:0051453 |
regulation of intracellular pH(GO:0051453) |
| 0.0 |
0.0 |
GO:0000019 |
regulation of mitotic recombination(GO:0000019) |
| 0.0 |
0.1 |
GO:0043525 |
positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.0 |
0.1 |
GO:0018101 |
protein citrullination(GO:0018101) |
| 0.0 |
0.2 |
GO:0046676 |
negative regulation of peptide secretion(GO:0002792) negative regulation of insulin secretion(GO:0046676) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.0 |
0.1 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 |
0.0 |
GO:0046851 |
negative regulation of bone resorption(GO:0045779) negative regulation of bone remodeling(GO:0046851) |
| 0.0 |
0.0 |
GO:0034036 |
purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 |
0.0 |
GO:0042074 |
cell migration involved in gastrulation(GO:0042074) |
| 0.0 |
0.3 |
GO:0006826 |
iron ion transport(GO:0006826) |
| 0.0 |
0.1 |
GO:0072676 |
lymphocyte chemotaxis(GO:0048247) lymphocyte migration(GO:0072676) |
| 0.0 |
0.4 |
GO:0031295 |
lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |