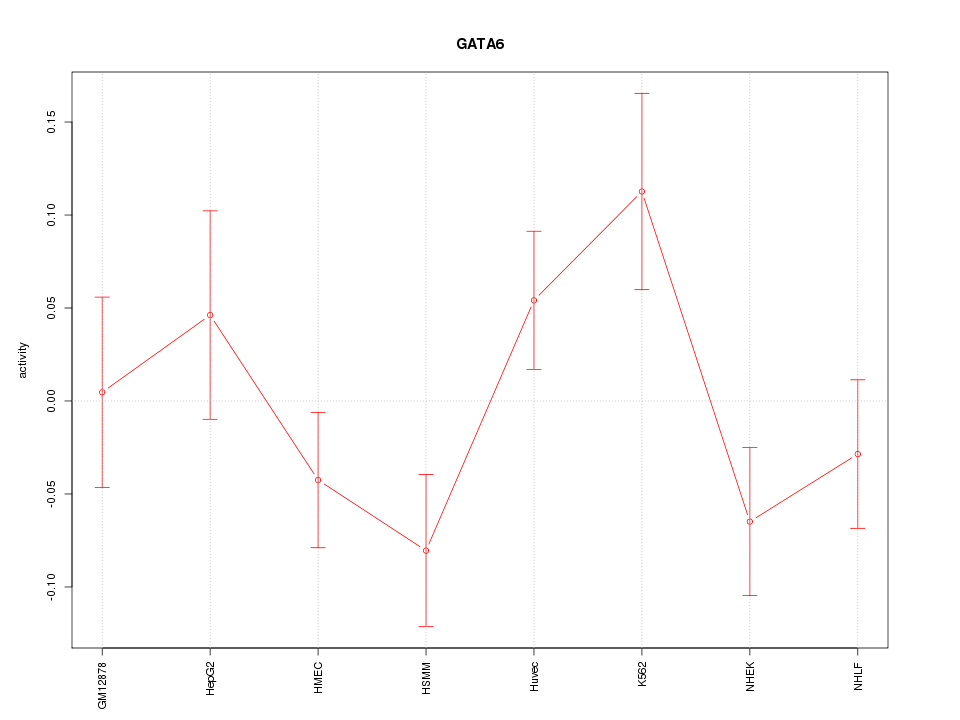
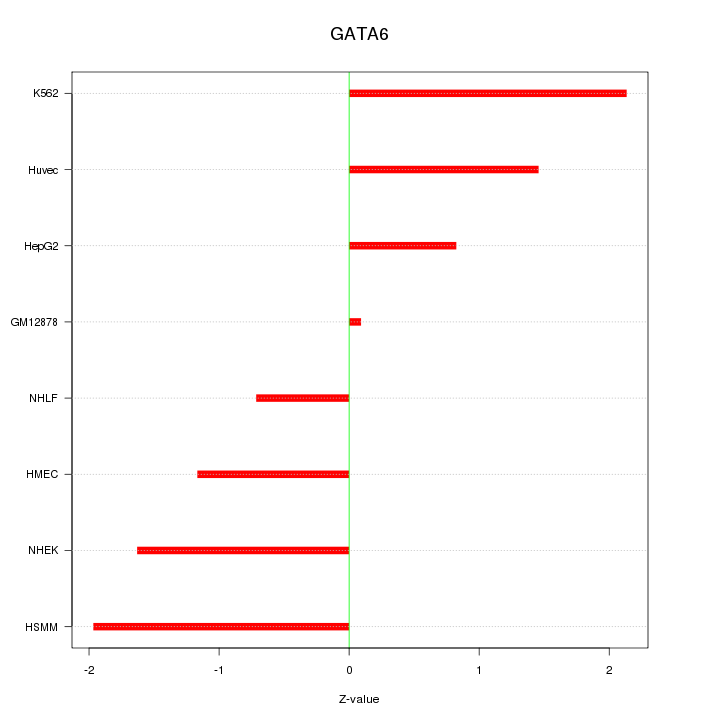
Motif ID: GATA6
Z-value: 1.404
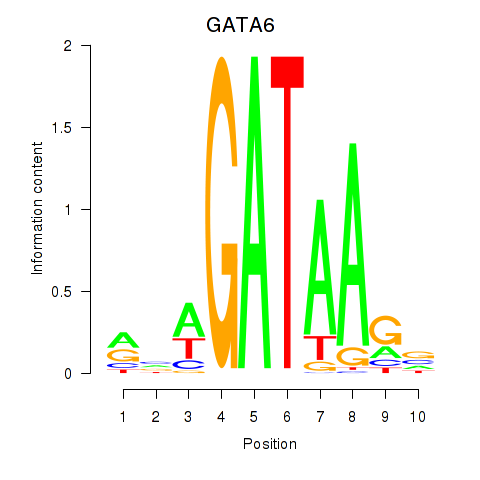
Transcription factors associated with GATA6:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| GATA6 | ENSG00000141448.4 | GATA6 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.6 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.5 | 1.5 | GO:0097237 | response to cobalt ion(GO:0032025) cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.5 | 3.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.4 | 2.6 | GO:1904729 | regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.3 | 1.0 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.2 | 3.2 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.2 | 0.7 | GO:0071635 | negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.1 | 0.4 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) positive regulation of sarcomere organization(GO:0060298) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.9 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.1 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 1.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.3 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.2 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:1903115 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.0 | 0.2 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 1.2 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.7 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 1.4 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
| 0.0 | 1.2 | GO:0050931 | melanocyte differentiation(GO:0030318) pigment cell differentiation(GO:0050931) |
| 0.0 | 2.0 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.2 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.8 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.4 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.2 | GO:0048066 | developmental pigmentation(GO:0048066) |
| 0.0 | 1.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.7 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 8.3 | GO:0016337 | single organismal cell-cell adhesion(GO:0016337) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.0 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 1.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 1.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 3.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 1.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.9 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) |
| 0.1 | 1.6 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 0.2 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 3.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 1.1 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 1.2 | GO:0005643 | nuclear pore(GO:0005643) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.4 | 1.6 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.3 | 0.9 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.3 | 3.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.2 | 1.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 2.4 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.2 | 1.0 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 1.5 | GO:0043176 | amine binding(GO:0043176) |
| 0.1 | 3.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 2.2 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 9.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.9 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.4 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.3 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.6 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.1 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 2.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.9 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | ST_INTERLEUKIN_4_PATHWAY | Interleukin 4 (IL-4) Pathway |