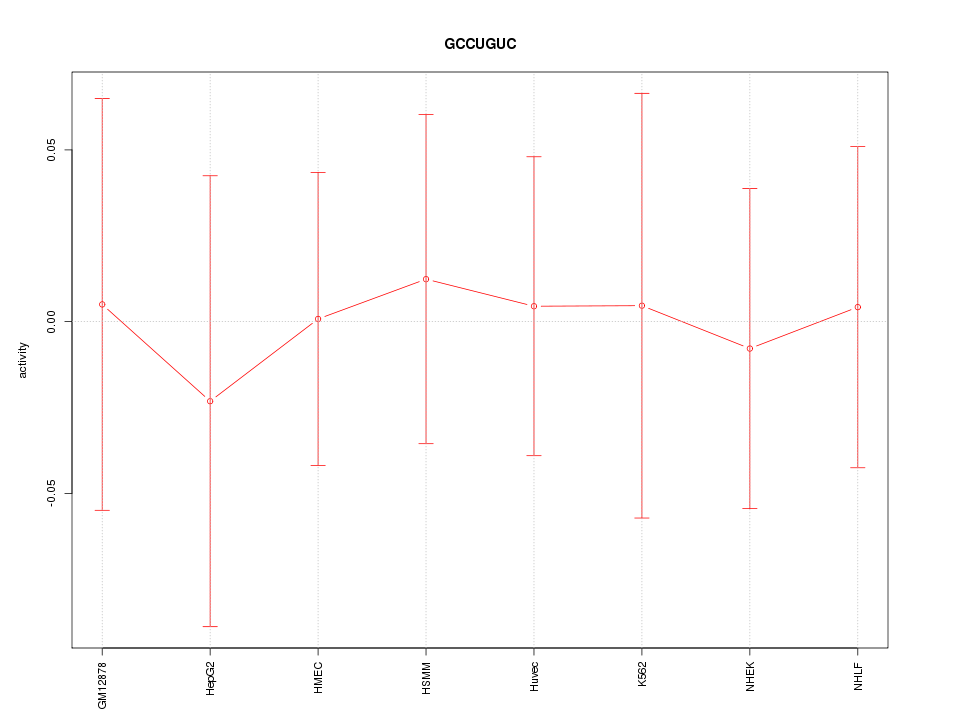
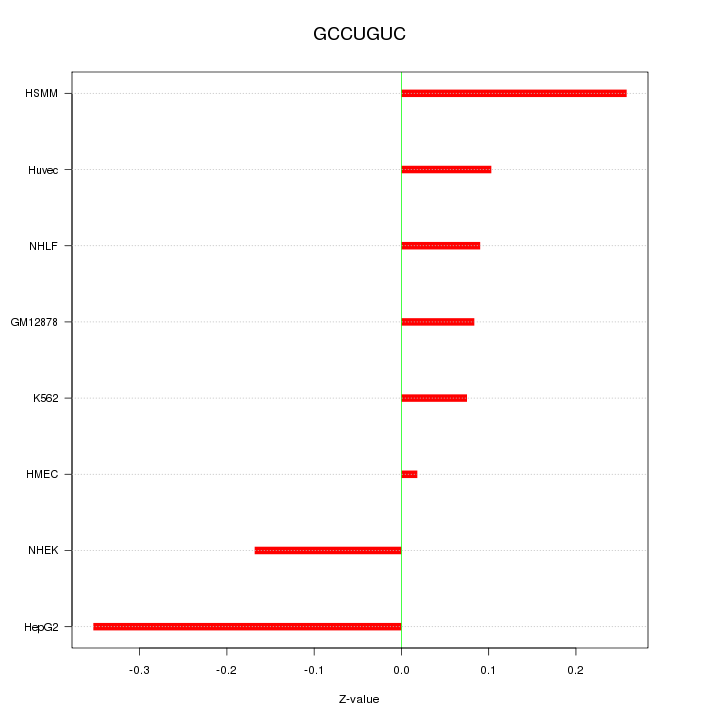
|
chr7_-_19157248
|
0.117
|
ENST00000242261.5
|
TWIST1
|
twist family bHLH transcription factor 1
|
|
chr1_-_244013384
|
0.067
|
ENST00000366539.1
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3
|
|
chr13_+_114238997
|
0.059
|
ENST00000538138.1
ENST00000375370.5
|
TFDP1
|
transcription factor Dp-1
|
|
chr3_+_152017181
|
0.056
|
ENST00000498502.1
ENST00000324196.5
ENST00000545754.1
ENST00000357472.3
|
MBNL1
|
muscleblind-like splicing regulator 1
|
|
chr1_+_145524891
|
0.054
|
ENST00000369304.3
|
ITGA10
|
integrin, alpha 10
|
|
chr4_+_55095264
|
0.047
|
ENST00000257290.5
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr5_-_146833485
|
0.042
|
ENST00000398514.3
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr2_-_217560248
|
0.042
|
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr20_-_23030296
|
0.039
|
ENST00000377103.2
|
THBD
|
thrombomodulin
|
|
chr3_-_171178157
|
0.038
|
ENST00000465393.1
ENST00000436636.2
ENST00000369326.5
ENST00000538048.1
ENST00000341852.6
|
TNIK
|
TRAF2 and NCK interacting kinase
|
|
chr3_-_114790179
|
0.038
|
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr6_+_44187242
|
0.036
|
ENST00000393844.1
|
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1
|
|
chr10_-_118032697
|
0.036
|
ENST00000439649.3
|
GFRA1
|
GDNF family receptor alpha 1
|
|
chr1_-_203055129
|
0.034
|
ENST00000241651.4
|
MYOG
|
myogenin (myogenic factor 4)
|
|
chr12_-_54673871
|
0.033
|
ENST00000209875.4
|
CBX5
|
chromobox homolog 5
|
|
chr11_-_6633799
|
0.031
|
ENST00000299424.4
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa
|
|
chr11_+_35684288
|
0.030
|
ENST00000299413.5
|
TRIM44
|
tripartite motif containing 44
|
|
chr19_+_30433110
|
0.029
|
ENST00000542441.2
ENST00000392271.1
|
URI1
|
URI1, prefoldin-like chaperone
|
|
chr17_-_1465924
|
0.029
|
ENST00000573231.1
ENST00000576722.1
ENST00000576761.1
ENST00000576010.2
ENST00000313486.7
ENST00000539476.1
|
PITPNA
|
phosphatidylinositol transfer protein, alpha
|
|
chr17_+_46184911
|
0.027
|
ENST00000580219.1
ENST00000452859.2
ENST00000393405.2
ENST00000439357.2
ENST00000359238.2
|
SNX11
|
sorting nexin 11
|
|
chr12_-_42632016
|
0.026
|
ENST00000442791.3
ENST00000327791.4
ENST00000534854.2
ENST00000380788.3
ENST00000380790.4
|
YAF2
|
YY1 associated factor 2
|
|
chr2_+_201676256
|
0.025
|
ENST00000452206.1
ENST00000410110.2
ENST00000409600.1
|
BZW1
|
basic leucine zipper and W2 domains 1
|
|
chr12_+_70760056
|
0.025
|
ENST00000258111.4
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chr15_-_89456593
|
0.024
|
ENST00000558029.1
ENST00000539437.1
ENST00000542878.1
ENST00000268151.7
ENST00000566497.1
|
MFGE8
|
milk fat globule-EGF factor 8 protein
|
|
chr16_-_85722530
|
0.024
|
ENST00000253462.3
|
GINS2
|
GINS complex subunit 2 (Psf2 homolog)
|
|
chr5_+_132387633
|
0.023
|
ENST00000304858.2
|
HSPA4
|
heat shock 70kDa protein 4
|
|
chr12_-_56583332
|
0.023
|
ENST00000347471.4
ENST00000267064.4
ENST00000394023.3
|
SMARCC2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2
|
|
chr6_-_75994536
|
0.022
|
ENST00000475111.2
ENST00000230461.6
|
TMEM30A
|
transmembrane protein 30A
|
|
chr6_+_4021554
|
0.021
|
ENST00000337659.6
|
PRPF4B
|
pre-mRNA processing factor 4B
|
|
chr9_+_77112244
|
0.020
|
ENST00000376896.3
|
RORB
|
RAR-related orphan receptor B
|
|
chr2_+_191273052
|
0.019
|
ENST00000417958.1
ENST00000432036.1
ENST00000392328.1
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr9_-_117568365
|
0.019
|
ENST00000374045.4
|
TNFSF15
|
tumor necrosis factor (ligand) superfamily, member 15
|
|
chr14_-_61116168
|
0.017
|
ENST00000247182.6
|
SIX1
|
SIX homeobox 1
|
|
chr3_-_125094093
|
0.016
|
ENST00000484491.1
ENST00000492394.1
ENST00000471196.1
ENST00000468369.1
ENST00000544464.1
ENST00000485866.1
ENST00000360647.4
|
ZNF148
|
zinc finger protein 148
|
|
chr22_-_42466782
|
0.016
|
ENST00000396398.3
ENST00000403363.1
ENST00000402937.1
|
NAGA
|
N-acetylgalactosaminidase, alpha-
|
|
chr11_-_6704513
|
0.015
|
ENST00000532203.1
ENST00000288937.6
|
MRPL17
|
mitochondrial ribosomal protein L17
|
|
chr20_+_34894247
|
0.014
|
ENST00000373913.3
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4
|
|
chr7_-_127983877
|
0.014
|
ENST00000415472.2
ENST00000478061.1
ENST00000223073.2
ENST00000459726.1
|
RBM28
|
RNA binding motif protein 28
|
|
chr1_+_245133062
|
0.014
|
ENST00000366523.1
|
EFCAB2
|
EF-hand calcium binding domain 2
|
|
chr17_-_45266542
|
0.014
|
ENST00000531206.1
ENST00000527547.1
ENST00000446365.2
ENST00000575483.1
ENST00000066544.3
|
CDC27
|
cell division cycle 27
|
|
chr6_+_44238203
|
0.014
|
ENST00000451188.2
|
TMEM151B
|
transmembrane protein 151B
|
|
chr17_+_30264014
|
0.014
|
ENST00000322652.5
ENST00000580398.1
|
SUZ12
|
SUZ12 polycomb repressive complex 2 subunit
|
|
chr14_+_75348592
|
0.013
|
ENST00000334220.4
|
DLST
|
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex)
|
|
chr10_-_43903217
|
0.012
|
ENST00000357065.4
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F
|
|
chr3_+_137906109
|
0.012
|
ENST00000481646.1
ENST00000469044.1
ENST00000491704.1
ENST00000461600.1
|
ARMC8
|
armadillo repeat containing 8
|
|
chr17_+_30677136
|
0.011
|
ENST00000394670.4
ENST00000321233.6
ENST00000394673.2
ENST00000341711.6
ENST00000579634.1
ENST00000580759.1
ENST00000342555.6
ENST00000577908.1
ENST00000394679.5
ENST00000582165.1
|
ZNF207
|
zinc finger protein 207
|
|
chr1_+_36348790
|
0.011
|
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1
|
|
chr19_-_15560730
|
0.010
|
ENST00000389282.4
ENST00000263381.7
|
WIZ
|
widely interspaced zinc finger motifs
|
|
chr11_-_6624801
|
0.010
|
ENST00000534343.1
ENST00000254605.6
|
RRP8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast)
|
|
chr9_-_74980113
|
0.010
|
ENST00000376962.5
ENST00000376960.4
ENST00000237937.3
|
ZFAND5
|
zinc finger, AN1-type domain 5
|
|
chr15_+_92937058
|
0.010
|
ENST00000268164.3
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2
|
|
chr1_-_23857698
|
0.010
|
ENST00000361729.2
|
E2F2
|
E2F transcription factor 2
|
|
chr8_+_59323823
|
0.009
|
ENST00000399598.2
|
UBXN2B
|
UBX domain protein 2B
|
|
chr17_-_61523535
|
0.008
|
ENST00000584031.1
ENST00000392976.1
|
CYB561
|
cytochrome b561
|
|
chr19_+_39616410
|
0.007
|
ENST00000602004.1
ENST00000599470.1
ENST00000321944.4
ENST00000593480.1
ENST00000358301.3
ENST00000593690.1
ENST00000599386.1
|
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4
|
|
chr16_-_69419871
|
0.007
|
ENST00000603068.1
ENST00000254942.3
ENST00000567296.2
|
TERF2
|
telomeric repeat binding factor 2
|
|
chr6_-_39197226
|
0.007
|
ENST00000359534.3
|
KCNK5
|
potassium channel, subfamily K, member 5
|
|
chr17_-_39890893
|
0.007
|
ENST00000393939.2
ENST00000347901.4
ENST00000341193.5
ENST00000310778.5
|
HAP1
|
huntingtin-associated protein 1
|
|
chr11_-_62572901
|
0.007
|
ENST00000439713.2
ENST00000531131.1
ENST00000530875.1
ENST00000531709.2
ENST00000294172.2
|
NXF1
|
nuclear RNA export factor 1
|
|
chrX_+_53078273
|
0.005
|
ENST00000332582.4
|
GPR173
|
G protein-coupled receptor 173
|
|
chr2_+_25015968
|
0.005
|
ENST00000380834.2
ENST00000473706.1
|
CENPO
|
centromere protein O
|
|
chr3_+_37493610
|
0.005
|
ENST00000264741.5
|
ITGA9
|
integrin, alpha 9
|
|
chr16_+_54964740
|
0.005
|
ENST00000394636.4
|
IRX5
|
iroquois homeobox 5
|
|
chrX_+_99839799
|
0.005
|
ENST00000373031.4
|
TNMD
|
tenomodulin
|
|
chr1_+_117602925
|
0.003
|
ENST00000369466.4
|
TTF2
|
transcription termination factor, RNA polymerase II
|
|
chr14_+_60715928
|
0.003
|
ENST00000395076.4
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A
|
|
chr11_-_82782861
|
0.003
|
ENST00000524635.1
ENST00000526205.1
ENST00000527633.1
ENST00000533486.1
ENST00000533276.2
|
RAB30
|
RAB30, member RAS oncogene family
|
|
chr3_-_9291063
|
0.003
|
ENST00000383836.3
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr18_-_18691739
|
0.003
|
ENST00000399799.2
|
ROCK1
|
Rho-associated, coiled-coil containing protein kinase 1
|
|
chr4_+_699537
|
0.003
|
ENST00000419774.1
ENST00000362003.5
ENST00000400151.2
ENST00000427463.1
ENST00000470161.2
|
PCGF3
|
polycomb group ring finger 3
|
|
chr3_-_197686847
|
0.002
|
ENST00000265239.6
|
IQCG
|
IQ motif containing G
|
|
chr11_+_48002076
|
0.002
|
ENST00000418331.2
ENST00000440289.2
|
PTPRJ
|
protein tyrosine phosphatase, receptor type, J
|
|
chr5_-_137090028
|
0.001
|
ENST00000314940.4
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr20_+_10199468
|
0.001
|
ENST00000254976.2
ENST00000304886.2
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr16_+_29789561
|
0.001
|
ENST00000400752.4
|
ZG16
|
zymogen granule protein 16
|
|
chr4_-_39460520
|
0.001
|
ENST00000503040.1
ENST00000504470.1
ENST00000508595.1
ENST00000295955.9
|
RPL9
|
ribosomal protein L9
|
|
chr5_-_66492562
|
0.000
|
ENST00000256447.4
|
CD180
|
CD180 molecule
|
|
chr19_+_1752372
|
0.000
|
ENST00000382349.4
|
ONECUT3
|
one cut homeobox 3
|