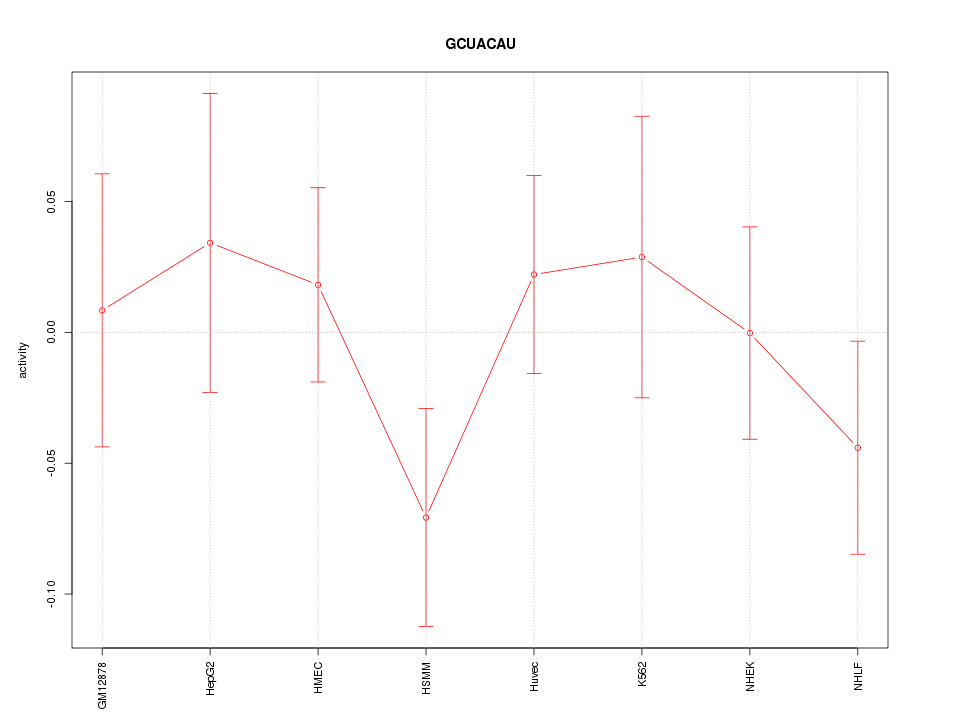
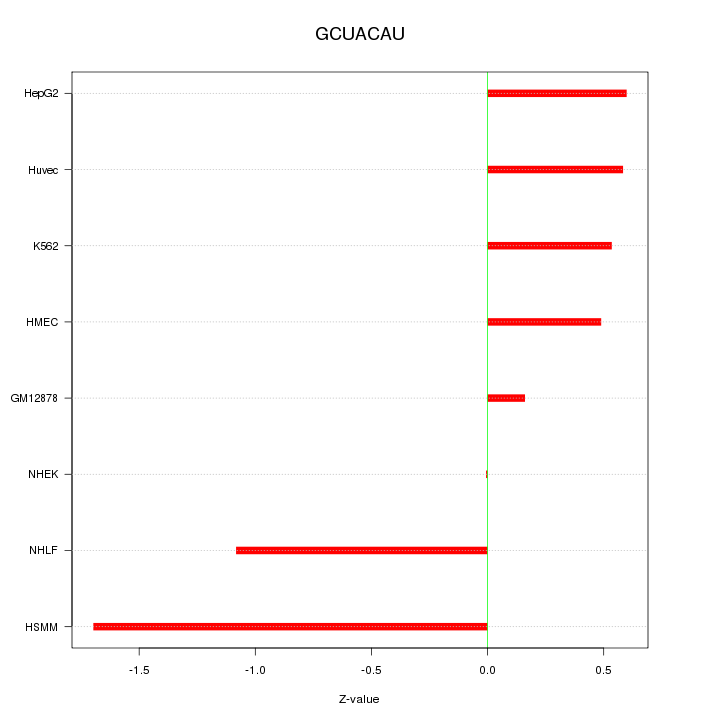
| 0.2 |
0.7 |
GO:0097237 |
autophagic cell death(GO:0048102) cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 |
0.4 |
GO:2000644 |
negative regulation of low-density lipoprotein particle clearance(GO:0010989) low-density lipoprotein particle receptor catabolic process(GO:0032802) regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.1 |
0.9 |
GO:0060123 |
regulation of growth hormone secretion(GO:0060123) |
| 0.1 |
0.3 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 |
0.1 |
GO:0043537 |
regulation of endothelial cell proliferation(GO:0001936) negative regulation of endothelial cell proliferation(GO:0001937) negative regulation of endothelial cell migration(GO:0010596) negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 |
0.3 |
GO:1901503 |
ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 |
0.3 |
GO:0035630 |
glycolate metabolic process(GO:0009441) bone mineralization involved in bone maturation(GO:0035630) |
| 0.0 |
0.1 |
GO:0003289 |
septum primum development(GO:0003284) atrial septum primum morphogenesis(GO:0003289) embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 |
0.2 |
GO:0060708 |
spongiotrophoblast differentiation(GO:0060708) |
| 0.0 |
0.3 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 |
0.2 |
GO:0006930 |
substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 |
0.2 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.0 |
0.2 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.0 |
0.1 |
GO:0046619 |
optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 |
0.1 |
GO:2000078 |
epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) regulation of type B pancreatic cell development(GO:2000074) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 |
0.2 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) positive regulation of adherens junction organization(GO:1903393) |
| 0.0 |
0.2 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 |
0.1 |
GO:0043619 |
regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 |
0.4 |
GO:0006386 |
transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 |
0.1 |
GO:0033152 |
immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 |
0.4 |
GO:0090630 |
activation of GTPase activity(GO:0090630) |
| 0.0 |
0.2 |
GO:0060487 |
lung epithelial cell differentiation(GO:0060487) |
| 0.0 |
0.1 |
GO:0060261 |
positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 |
0.2 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.0 |
0.1 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 |
0.1 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 |
0.0 |
GO:0008050 |
female courtship behavior(GO:0008050) |
| 0.0 |
1.8 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.2 |
GO:0001573 |
ganglioside metabolic process(GO:0001573) |
| 0.0 |
0.1 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |