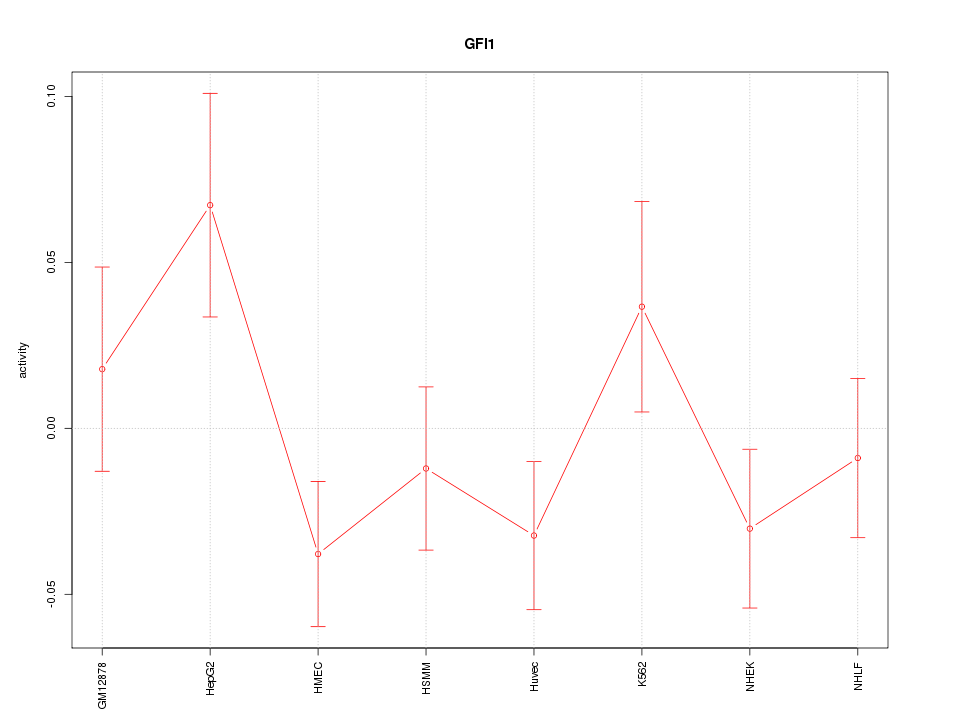
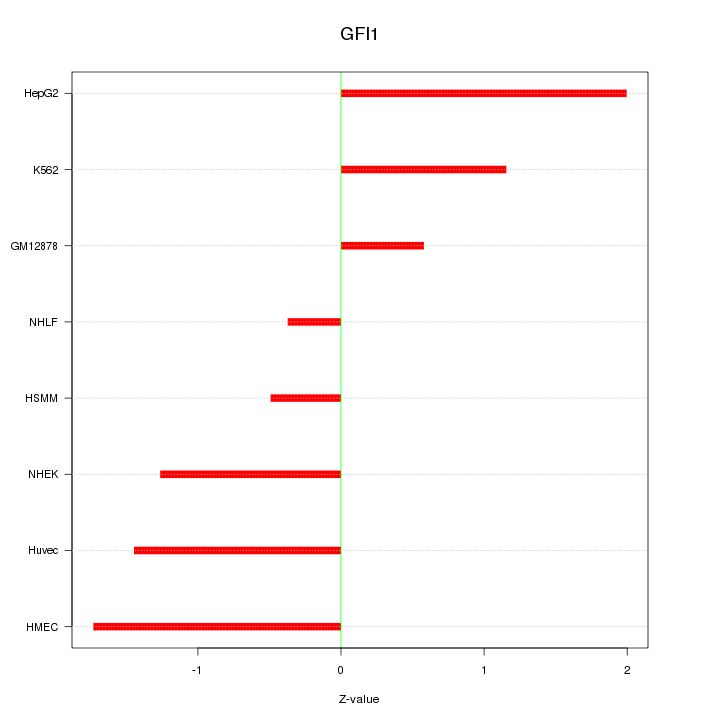
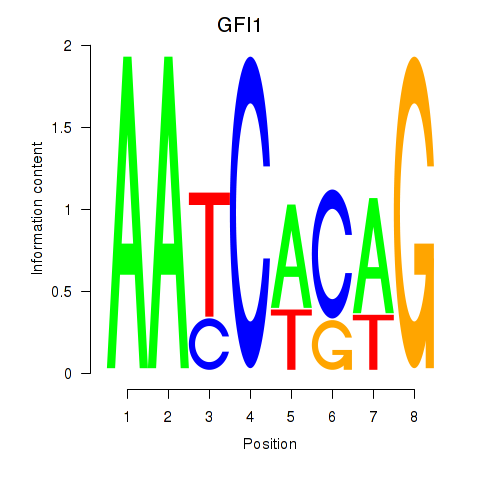
Motif ID: GFI1
Z-value: 1.261
Transcription factors associated with GFI1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| GFI1 | ENSG00000162676.7 | GFI1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.6 | 2.9 | GO:0007614 | short-term memory(GO:0007614) |
| 0.3 | 1.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.2 | 2.9 | GO:0055069 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.2 | 1.7 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.2 | 0.8 | GO:0031643 | regulation of Schwann cell differentiation(GO:0014038) positive regulation of Schwann cell differentiation(GO:0014040) positive regulation of myelination(GO:0031643) |
| 0.2 | 0.6 | GO:0090034 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.2 | 0.7 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 0.7 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.2 | 0.5 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.2 | 0.5 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 0.7 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.2 | 1.1 | GO:0019264 | L-serine catabolic process(GO:0006565) glycine biosynthetic process from serine(GO:0019264) |
| 0.2 | 0.6 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) actin filament reorganization(GO:0090527) |
| 0.1 | 0.7 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.1 | 0.4 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 0.6 | GO:0042905 | retinal metabolic process(GO:0042574) 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.6 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 0.4 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.1 | 0.4 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) regulation of meiosis I(GO:0060631) |
| 0.1 | 0.4 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 0.7 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.2 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 0.8 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 0.4 | GO:0071025 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.1 | 0.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.5 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.1 | 0.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.1 | 0.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.4 | GO:0051140 | tyrosine phosphorylation of Stat4 protein(GO:0042504) regulation of tyrosine phosphorylation of Stat4 protein(GO:0042519) positive regulation of tyrosine phosphorylation of Stat4 protein(GO:0042520) regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.1 | 0.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 4.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.7 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.3 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.1 | 0.2 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.1 | 0.2 | GO:0060209 | estrus(GO:0060209) |
| 0.1 | 0.8 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.5 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 1.5 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.2 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.3 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.0 | 0.2 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.0 | 0.1 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 | 0.5 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.2 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.2 | GO:0010759 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.9 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.4 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.5 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 2.7 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.4 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 1.6 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.3 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.5 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.3 | GO:0060487 | lung epithelial cell differentiation(GO:0060487) |
| 0.0 | 1.3 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 0.1 | GO:0061198 | fungiform papilla development(GO:0061196) fungiform papilla morphogenesis(GO:0061197) fungiform papilla formation(GO:0061198) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.2 | GO:0003148 | outflow tract septum morphogenesis(GO:0003148) |
| 0.0 | 2.1 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.0 | 0.9 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.2 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 0.7 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) hyperosmotic salinity response(GO:0042538) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.2 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.1 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 1.9 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.4 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.7 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
| 0.0 | 0.1 | GO:0032817 | regulation of natural killer cell proliferation(GO:0032817) |
| 0.0 | 0.4 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.0 | GO:0048865 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 | 0.0 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.9 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.1 | GO:0003097 | renal water transport(GO:0003097) renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.3 | GO:0034612 | response to tumor necrosis factor(GO:0034612) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0001660 | fever generation(GO:0001660) |
| 0.0 | 0.1 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.1 | GO:0000090 | mitotic anaphase(GO:0000090) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.2 | 2.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.4 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 0.4 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.1 | 0.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 2.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.8 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.5 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 1.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 3.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.4 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.8 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.6 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 2.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 1.1 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.3 | 1.7 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.3 | 2.8 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.2 | 0.9 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.2 | 0.9 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.2 | 0.8 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.2 | 2.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 0.5 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 1.1 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.2 | 0.5 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 0.6 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.4 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.1 | 0.4 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.7 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 2.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.5 | GO:0043499 | obsolete eukaryotic cell surface binding(GO:0043499) |
| 0.1 | 1.5 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.8 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.4 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 1.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.1 | 0.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.6 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.3 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.1 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.5 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.1 | 0.4 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.1 | 0.4 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.1 | 1.0 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.3 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.4 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.0 | 0.1 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.0 | 0.3 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.1 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.0 | 0.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 5.4 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.0 | GO:0034594 | phosphatidylinositol trisphosphate phosphatase activity(GO:0034594) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.6 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.1 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA_FAS_SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.0 | 0.5 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |