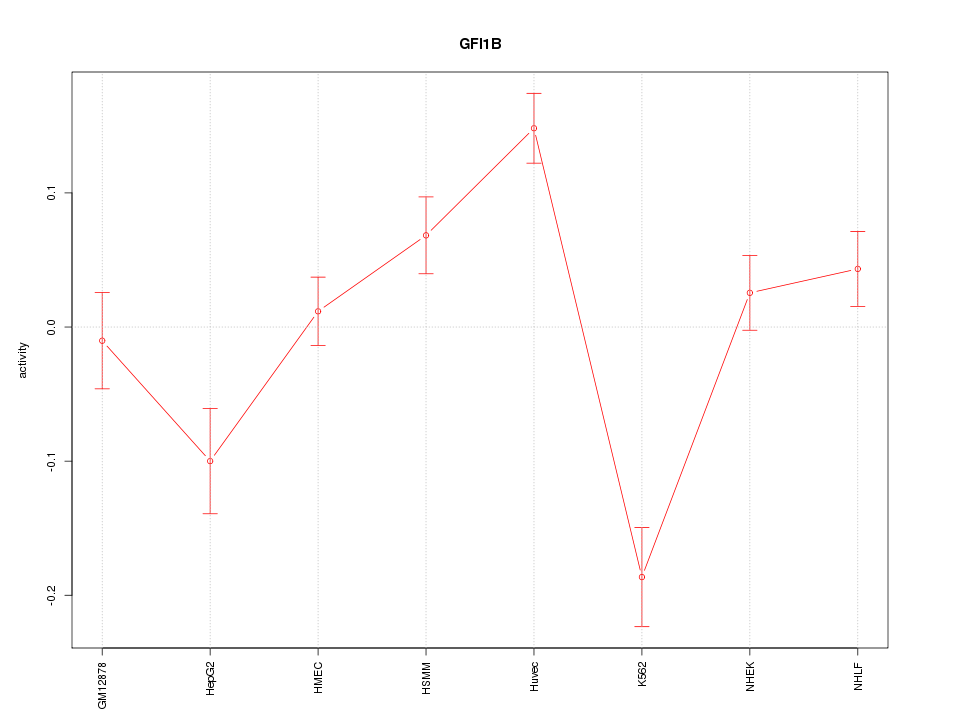
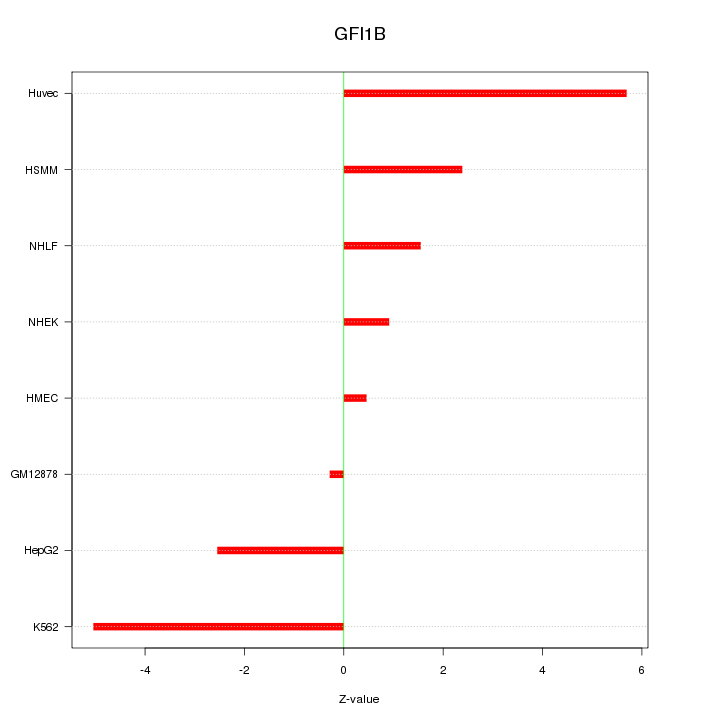
Motif ID: GFI1B
Z-value: 3.032
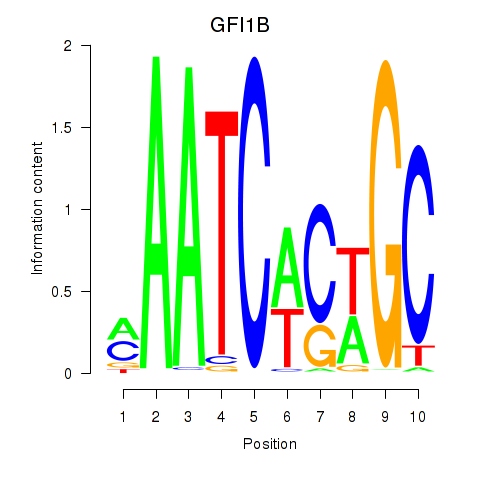
Transcription factors associated with GFI1B:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| GFI1B | ENSG00000165702.8 | GFI1B |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 1.6 | 6.5 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 1.2 | 3.5 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) dermatan sulfate biosynthetic process(GO:0030208) |
| 1.1 | 2.3 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.8 | 4.9 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.7 | 2.9 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.7 | 3.4 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.6 | 1.7 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.6 | 2.2 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.5 | 5.5 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.4 | 1.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.4 | 10.4 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.4 | 4.0 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.4 | 2.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.4 | 2.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.3 | 1.0 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.3 | 1.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.3 | 0.9 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.3 | 1.8 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.3 | 0.9 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.3 | 2.8 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.3 | 2.0 | GO:0006553 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.2 | 2.0 | GO:0030903 | notochord development(GO:0030903) |
| 0.2 | 3.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.2 | 1.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 1.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.2 | 0.6 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.2 | 5.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.2 | 1.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 2.0 | GO:0048488 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.2 | 1.8 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.1 | 0.9 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 6.4 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.1 | 1.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 12.1 | GO:0008064 | regulation of actin polymerization or depolymerization(GO:0008064) |
| 0.1 | 3.1 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.3 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.1 | 0.3 | GO:0060267 | respiratory burst involved in defense response(GO:0002679) lipopolysaccharide transport(GO:0015920) positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.4 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.1 | 0.6 | GO:0002005 | angiotensin catabolic process in blood(GO:0002005) |
| 0.1 | 0.3 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.6 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 2.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 0.3 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.1 | 0.4 | GO:0045080 | positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.1 | 1.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.2 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.1 | 1.3 | GO:1901224 | activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.1 | 0.4 | GO:0001507 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.1 | 0.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 1.1 | GO:0007128 | meiotic prophase I(GO:0007128) |
| 0.1 | 3.8 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 0.3 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.1 | 1.2 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 1.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 4.9 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 0.2 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 1.0 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.1 | 0.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 1.0 | GO:0050807 | regulation of synapse organization(GO:0050807) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.4 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 1.1 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.9 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.9 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.0 | 0.1 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 1.7 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.4 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.2 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 0.5 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 1.3 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 1.3 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 4.2 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 1.1 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 1.8 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.2 | GO:0060119 | auditory receptor cell development(GO:0060117) inner ear receptor cell development(GO:0060119) |
| 0.0 | 0.2 | GO:0046051 | GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.0 | 0.2 | GO:0045329 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 | 7.8 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 1.1 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.7 | GO:0042384 | cilium assembly(GO:0042384) |
| 0.0 | 2.6 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.9 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.9 | GO:0016477 | cell migration(GO:0016477) |
| 0.0 | 0.3 | GO:0044241 | lipid digestion(GO:0044241) |
| 0.0 | 0.2 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.0 | 0.1 | GO:0043249 | erythrocyte maturation(GO:0043249) enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.2 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.8 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.0 | 1.7 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.0 | 0.2 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 1.6 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.8 | 5.3 | GO:1990752 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.7 | 4.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.6 | 12.1 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.5 | 2.7 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.4 | 2.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.4 | 1.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 1.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 0.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 10.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 1.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 2.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.0 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 4.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.9 | GO:0099568 | cell cortex(GO:0005938) cytoplasmic region(GO:0099568) |
| 0.1 | 15.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 1.9 | GO:0032153 | cell division site(GO:0032153) cell division site part(GO:0032155) |
| 0.1 | 0.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 1.3 | GO:0043205 | fibril(GO:0043205) |
| 0.1 | 0.9 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.1 | 1.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 4.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.6 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 2.5 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 2.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.3 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 4.7 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.8 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.0 | 5.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 3.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.0 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 10.2 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 7.0 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 2.1 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 3.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 2.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 12.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.4 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.0 | 2.8 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0005912 | adherens junction(GO:0005912) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.1 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.7 | 4.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.6 | 3.7 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.6 | 1.8 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.5 | 12.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.4 | 2.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.4 | 2.7 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.3 | 1.0 | GO:0048184 | obsolete follistatin binding(GO:0048184) |
| 0.3 | 1.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.3 | 1.0 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.3 | 3.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.3 | 11.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.3 | 5.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 0.9 | GO:0033265 | acetylcholinesterase activity(GO:0003990) choline binding(GO:0033265) |
| 0.2 | 1.3 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.2 | 2.9 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.2 | 0.8 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 5.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 0.6 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.2 | 4.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 1.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.9 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 2.0 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 6.5 | GO:1990782 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.3 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) glutamate binding(GO:0016595) |
| 0.1 | 0.5 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.1 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 0.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.4 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 2.9 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.3 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.1 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 1.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 2.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.1 | 0.6 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 8.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.7 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.1 | 4.2 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 1.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 5.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.2 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.3 | GO:0046980 | oligopeptide-transporting ATPase activity(GO:0015421) tapasin binding(GO:0046980) |
| 0.0 | 0.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 1.1 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.8 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 1.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 6.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 6.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.1 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.9 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 2.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 9.3 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 3.5 | GO:0030528 | obsolete transcription regulator activity(GO:0030528) |
| 0.0 | 3.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.7 | GO:0016564 | obsolete transcription repressor activity(GO:0016564) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 13.6 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 4.8 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.5 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.3 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.6 | SA_FAS_SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 2.4 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.9 | SIG_BCR_SIGNALING_PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.8 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.6 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.7 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |