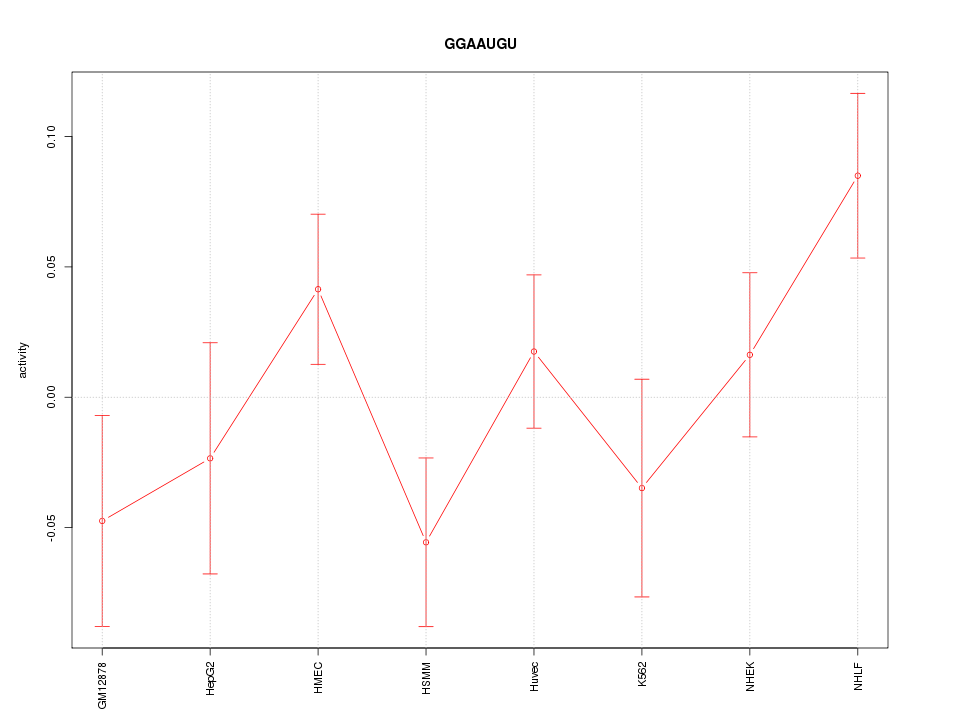
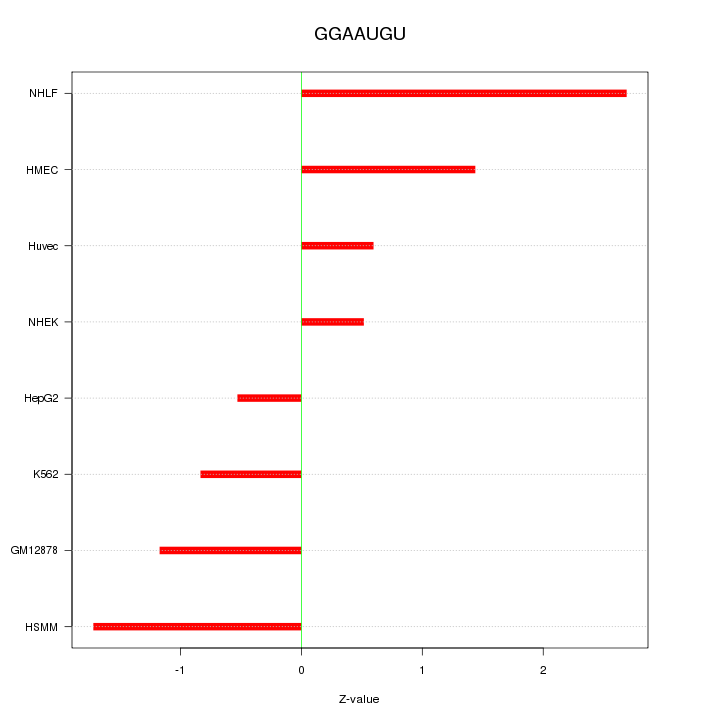
| 1.3 |
3.9 |
GO:0014029 |
neural crest formation(GO:0014029) neural crest cell fate commitment(GO:0014034) convergent extension involved in axis elongation(GO:0060028) cellular response to prostaglandin E stimulus(GO:0071380) response to heparin(GO:0071503) cellular response to heparin(GO:0071504) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.7 |
3.0 |
GO:0051503 |
purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) |
| 0.5 |
1.6 |
GO:0010757 |
negative regulation of plasminogen activation(GO:0010757) |
| 0.4 |
1.3 |
GO:0030205 |
dermatan sulfate metabolic process(GO:0030205) dermatan sulfate biosynthetic process(GO:0030208) |
| 0.3 |
2.0 |
GO:0060414 |
aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 |
0.8 |
GO:0006740 |
NADPH regeneration(GO:0006740) |
| 0.2 |
0.7 |
GO:0060054 |
positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.2 |
1.4 |
GO:0007499 |
ectoderm and mesoderm interaction(GO:0007499) |
| 0.2 |
1.0 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 |
0.6 |
GO:0042759 |
fatty acid elongation, saturated fatty acid(GO:0019367) long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 |
0.2 |
GO:0060024 |
rhythmic synaptic transmission(GO:0060024) |
| 0.2 |
0.5 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 |
1.0 |
GO:0007500 |
mesodermal cell fate determination(GO:0007500) |
| 0.2 |
1.4 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 |
1.3 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 |
1.8 |
GO:0050872 |
white fat cell differentiation(GO:0050872) |
| 0.1 |
0.8 |
GO:0043268 |
positive regulation of potassium ion transport(GO:0043268) |
| 0.1 |
1.4 |
GO:0001945 |
lymph vessel development(GO:0001945) |
| 0.1 |
0.8 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 |
0.4 |
GO:0031340 |
positive regulation of vesicle fusion(GO:0031340) |
| 0.1 |
0.3 |
GO:0001757 |
somite specification(GO:0001757) |
| 0.1 |
0.4 |
GO:0033197 |
response to vitamin E(GO:0033197) |
| 0.1 |
0.6 |
GO:0006880 |
intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 |
0.2 |
GO:0034770 |
histone H4-K20 methylation(GO:0034770) |
| 0.1 |
0.3 |
GO:0051409 |
response to nitrosative stress(GO:0051409) |
| 0.1 |
0.2 |
GO:0005997 |
xylulose metabolic process(GO:0005997) |
| 0.1 |
0.5 |
GO:0008354 |
germ cell migration(GO:0008354) |
| 0.1 |
0.2 |
GO:0018377 |
N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.1 |
0.4 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.1 |
0.2 |
GO:0046102 |
nicotinamide riboside catabolic process(GO:0006738) inosine metabolic process(GO:0046102) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.1 |
0.3 |
GO:0006930 |
substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.1 |
0.5 |
GO:0046835 |
carbohydrate phosphorylation(GO:0046835) |
| 0.1 |
0.4 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.0 |
0.2 |
GO:0021527 |
spinal cord association neuron differentiation(GO:0021527) |
| 0.0 |
0.1 |
GO:0007538 |
primary sex determination(GO:0007538) |
| 0.0 |
0.3 |
GO:0006477 |
protein sulfation(GO:0006477) |
| 0.0 |
0.1 |
GO:0060318 |
definitive erythrocyte differentiation(GO:0060318) |
| 0.0 |
0.3 |
GO:0046325 |
negative regulation of glucose import(GO:0046325) |
| 0.0 |
0.2 |
GO:0043116 |
negative regulation of vascular permeability(GO:0043116) |
| 0.0 |
0.1 |
GO:0032119 |
sequestering of zinc ion(GO:0032119) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 |
0.2 |
GO:0034204 |
Cdc42 protein signal transduction(GO:0032488) lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 |
0.5 |
GO:0034405 |
response to fluid shear stress(GO:0034405) |
| 0.0 |
0.1 |
GO:1904358 |
positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 |
0.3 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 |
0.1 |
GO:0010508 |
positive regulation of autophagy(GO:0010508) |
| 0.0 |
0.1 |
GO:0061298 |
progesterone secretion(GO:0042701) retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 |
0.1 |
GO:0046469 |
layer formation in cerebral cortex(GO:0021819) platelet activating factor metabolic process(GO:0046469) |
| 0.0 |
0.0 |
GO:0097576 |
autophagosome maturation(GO:0097352) vacuole fusion(GO:0097576) |
| 0.0 |
0.2 |
GO:0002553 |
histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 |
0.3 |
GO:0043113 |
receptor clustering(GO:0043113) |
| 0.0 |
0.3 |
GO:0045047 |
protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 |
0.6 |
GO:0048009 |
insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 |
0.2 |
GO:0060158 |
neuron remodeling(GO:0016322) regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 |
0.1 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.0 |
0.1 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) |
| 0.0 |
0.1 |
GO:0051927 |
obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.0 |
0.2 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 |
0.5 |
GO:0015813 |
L-glutamate transport(GO:0015813) |
| 0.0 |
0.1 |
GO:0035434 |
copper ion transmembrane transport(GO:0035434) |
| 0.0 |
0.4 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
0.4 |
GO:0006829 |
zinc II ion transport(GO:0006829) |
| 0.0 |
0.1 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) |
| 0.0 |
0.0 |
GO:0010996 |
mechanosensory behavior(GO:0007638) response to auditory stimulus(GO:0010996) operant conditioning(GO:0035106) |
| 0.0 |
0.3 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
0.4 |
GO:0034199 |
activation of protein kinase A activity(GO:0034199) |
| 0.0 |
1.3 |
GO:0008064 |
regulation of actin polymerization or depolymerization(GO:0008064) |
| 0.0 |
0.3 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.0 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 |
0.0 |
GO:0010544 |
regulation of phosphatidylinositol biosynthetic process(GO:0010511) negative regulation of platelet activation(GO:0010544) regulation of phospholipid biosynthetic process(GO:0071071) |
| 0.0 |
0.1 |
GO:0006906 |
vesicle fusion(GO:0006906) |
| 0.0 |
0.1 |
GO:0061339 |
establishment of apical/basal cell polarity(GO:0035089) establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 |
0.9 |
GO:0006939 |
smooth muscle contraction(GO:0006939) |
| 0.0 |
0.2 |
GO:0030201 |
heparan sulfate proteoglycan metabolic process(GO:0030201) |
| 0.0 |
0.1 |
GO:0046519 |
sphingoid metabolic process(GO:0046519) |
| 0.0 |
0.0 |
GO:0060124 |
positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 |
0.7 |
GO:0031668 |
cellular response to extracellular stimulus(GO:0031668) |
| 0.0 |
0.3 |
GO:0080171 |
lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |