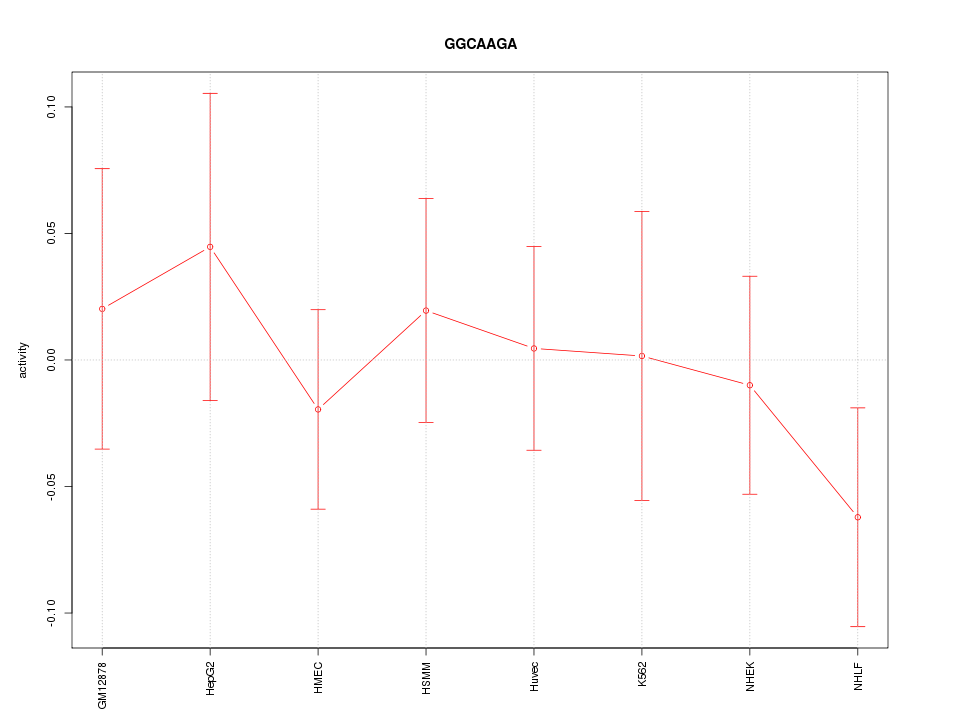
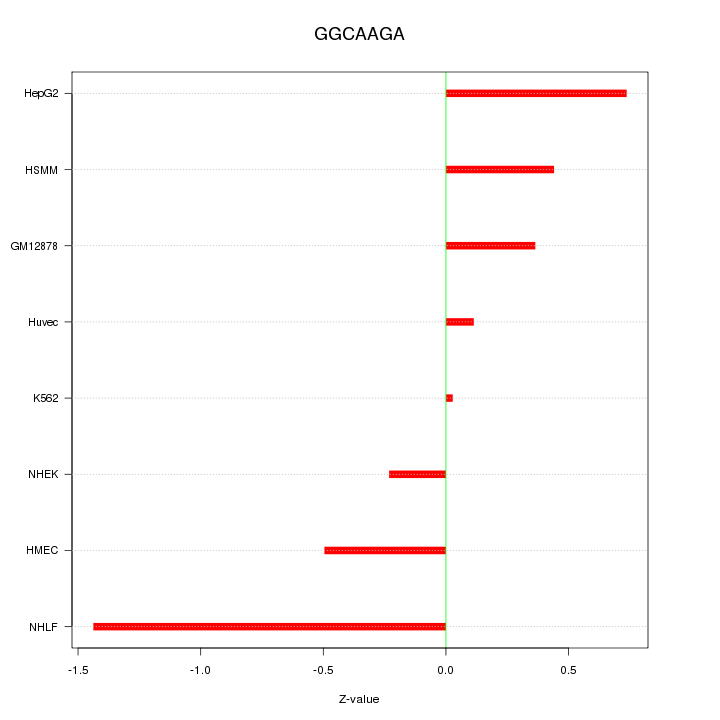
|
chr8_+_61591337
|
0.699
|
ENST00000423902.2
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr16_+_69599861
|
0.496
|
ENST00000354436.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr1_+_65613217
|
0.492
|
ENST00000545314.1
|
AK4
|
adenylate kinase 4
|
|
chr5_-_78809950
|
0.351
|
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila)
|
|
chr10_-_116286656
|
0.323
|
ENST00000428430.1
ENST00000369266.3
ENST00000392952.3
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr2_-_64881018
|
0.319
|
ENST00000313349.3
|
SERTAD2
|
SERTA domain containing 2
|
|
chr2_+_7057523
|
0.305
|
ENST00000320892.6
|
RNF144A
|
ring finger protein 144A
|
|
chr11_-_86666427
|
0.281
|
ENST00000531380.1
|
FZD4
|
frizzled family receptor 4
|
|
chr20_+_35974532
|
0.267
|
ENST00000373578.2
|
SRC
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog
|
|
chr17_-_65241281
|
0.256
|
ENST00000358691.5
ENST00000580168.1
|
HELZ
|
helicase with zinc finger
|
|
chr16_+_70557685
|
0.247
|
ENST00000302516.5
ENST00000566095.2
ENST00000577085.1
ENST00000567654.1
|
SF3B3
|
splicing factor 3b, subunit 3, 130kDa
|
|
chr19_-_33793430
|
0.238
|
ENST00000498907.2
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha
|
|
chr1_-_114355083
|
0.236
|
ENST00000261441.5
|
RSBN1
|
round spermatid basic protein 1
|
|
chr6_+_7107999
|
0.226
|
ENST00000491191.1
ENST00000379938.2
ENST00000471433.1
|
RREB1
|
ras responsive element binding protein 1
|
|
chr1_-_154842741
|
0.223
|
ENST00000271915.4
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr10_-_62704005
|
0.205
|
ENST00000337910.5
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr5_-_168006591
|
0.199
|
ENST00000239231.6
|
PANK3
|
pantothenate kinase 3
|
|
chr9_+_470288
|
0.189
|
ENST00000382303.1
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr2_+_61404624
|
0.184
|
ENST00000394457.3
|
AHSA2
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast)
|
|
chr11_-_35440796
|
0.178
|
ENST00000278379.3
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr12_-_31479045
|
0.177
|
ENST00000539409.1
ENST00000395766.1
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chrX_-_33146477
|
0.165
|
ENST00000378677.2
|
DMD
|
dystrophin
|
|
chr1_+_27022485
|
0.163
|
ENST00000324856.7
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr1_+_27153173
|
0.163
|
ENST00000374142.4
|
ZDHHC18
|
zinc finger, DHHC-type containing 18
|
|
chr12_-_42538657
|
0.162
|
ENST00000398675.3
|
GXYLT1
|
glucoside xylosyltransferase 1
|
|
chr3_-_114790179
|
0.158
|
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr7_+_94139105
|
0.151
|
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1
|
|
chr1_+_110527308
|
0.151
|
ENST00000369799.5
|
AHCYL1
|
adenosylhomocysteinase-like 1
|
|
chr17_+_64961026
|
0.149
|
ENST00000262138.3
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4
|
|
chr1_-_78444776
|
0.148
|
ENST00000370767.1
ENST00000421641.1
|
FUBP1
|
far upstream element (FUSE) binding protein 1
|
|
chr14_-_34420259
|
0.144
|
ENST00000250457.3
ENST00000547327.2
|
EGLN3
|
egl-9 family hypoxia-inducible factor 3
|
|
chr5_+_174905398
|
0.140
|
ENST00000321442.5
|
SFXN1
|
sideroflexin 1
|
|
chr4_+_87856129
|
0.130
|
ENST00000395146.4
ENST00000507468.1
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr18_+_55711575
|
0.130
|
ENST00000356462.6
ENST00000400345.3
ENST00000589054.1
ENST00000256832.7
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase
|
|
chr15_-_71055878
|
0.125
|
ENST00000322954.6
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr2_+_120770645
|
0.124
|
ENST00000443902.2
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5
|
|
chr1_+_226411319
|
0.122
|
ENST00000542034.1
ENST00000366810.5
|
MIXL1
|
Mix paired-like homeobox
|
|
chr12_-_49110613
|
0.121
|
ENST00000261900.3
|
CCNT1
|
cyclin T1
|
|
chr1_-_108507631
|
0.120
|
ENST00000527011.1
ENST00000370056.4
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr1_-_231560790
|
0.119
|
ENST00000366641.3
|
EGLN1
|
egl-9 family hypoxia-inducible factor 1
|
|
chr1_+_70876891
|
0.118
|
ENST00000411986.2
|
CTH
|
cystathionase (cystathionine gamma-lyase)
|
|
chr3_+_139654018
|
0.114
|
ENST00000458420.3
|
CLSTN2
|
calsyntenin 2
|
|
chr17_-_79885576
|
0.113
|
ENST00000574686.1
ENST00000357736.4
|
MAFG
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G
|
|
chr1_+_167190066
|
0.110
|
ENST00000367866.2
ENST00000429375.2
ENST00000452019.1
ENST00000420254.3
ENST00000541643.3
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr10_-_126432619
|
0.108
|
ENST00000337318.3
|
FAM53B
|
family with sequence similarity 53, member B
|
|
chr8_-_81083731
|
0.107
|
ENST00000379096.5
|
TPD52
|
tumor protein D52
|
|
chr20_+_3190006
|
0.106
|
ENST00000380113.3
ENST00000455664.2
ENST00000399838.3
|
ITPA
|
inosine triphosphatase (nucleoside triphosphate pyrophosphatase)
|
|
chr2_-_203776864
|
0.105
|
ENST00000261015.4
|
WDR12
|
WD repeat domain 12
|
|
chr4_+_184020398
|
0.104
|
ENST00000403733.3
ENST00000378925.3
|
WWC2
|
WW and C2 domain containing 2
|
|
chr4_+_106629929
|
0.102
|
ENST00000512828.1
ENST00000394730.3
ENST00000507281.1
ENST00000515279.1
|
GSTCD
|
glutathione S-transferase, C-terminal domain containing
|
|
chr2_-_200322723
|
0.102
|
ENST00000417098.1
|
SATB2
|
SATB homeobox 2
|
|
chr5_-_59189545
|
0.097
|
ENST00000340635.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_+_7342178
|
0.096
|
ENST00000266563.5
ENST00000543974.1
|
PEX5
|
peroxisomal biogenesis factor 5
|
|
chr12_-_498620
|
0.096
|
ENST00000399788.2
ENST00000382815.4
|
KDM5A
|
lysine (K)-specific demethylase 5A
|
|
chr16_-_70472946
|
0.096
|
ENST00000342907.2
|
ST3GAL2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2
|
|
chr4_-_42659102
|
0.095
|
ENST00000264449.10
ENST00000510289.1
ENST00000381668.5
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr3_-_185216766
|
0.094
|
ENST00000296254.3
|
TMEM41A
|
transmembrane protein 41A
|
|
chr17_-_1532106
|
0.091
|
ENST00000301335.5
ENST00000382147.4
|
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2
|
|
chr19_-_44100275
|
0.090
|
ENST00000422989.1
ENST00000598324.1
|
IRGQ
|
immunity-related GTPase family, Q
|
|
chr7_-_35734730
|
0.086
|
ENST00000396081.1
ENST00000311350.3
|
HERPUD2
|
HERPUD family member 2
|
|
chr16_-_402639
|
0.085
|
ENST00000262320.3
|
AXIN1
|
axin 1
|
|
chr11_-_72853091
|
0.082
|
ENST00000311172.7
ENST00000409314.1
|
FCHSD2
|
FCH and double SH3 domains 2
|
|
chr3_+_180630090
|
0.081
|
ENST00000357559.4
ENST00000305586.7
|
FXR1
|
fragile X mental retardation, autosomal homolog 1
|
|
chr20_+_60718785
|
0.080
|
ENST00000421564.1
ENST00000450482.1
ENST00000331758.3
|
SS18L1
|
synovial sarcoma translocation gene on chromosome 18-like 1
|
|
chr6_-_99395787
|
0.080
|
ENST00000369244.2
ENST00000229971.1
|
FBXL4
|
F-box and leucine-rich repeat protein 4
|
|
chr4_+_38665810
|
0.079
|
ENST00000261438.5
ENST00000514033.1
|
KLF3
|
Kruppel-like factor 3 (basic)
|
|
chr3_-_133614597
|
0.078
|
ENST00000285208.4
ENST00000460865.3
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr4_-_90229142
|
0.078
|
ENST00000609438.1
|
GPRIN3
|
GPRIN family member 3
|
|
chr12_+_862089
|
0.077
|
ENST00000315939.6
ENST00000537687.1
ENST00000447667.2
|
WNK1
|
WNK lysine deficient protein kinase 1
|
|
chr11_+_119076745
|
0.076
|
ENST00000264033.4
|
CBL
|
Cbl proto-oncogene, E3 ubiquitin protein ligase
|
|
chr1_+_161736072
|
0.076
|
ENST00000367942.3
|
ATF6
|
activating transcription factor 6
|
|
chr13_-_72441315
|
0.076
|
ENST00000305425.4
ENST00000313174.7
ENST00000354591.4
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr4_-_41216619
|
0.076
|
ENST00000508676.1
ENST00000506352.1
ENST00000295974.8
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr9_-_37034028
|
0.075
|
ENST00000520281.1
ENST00000446742.1
ENST00000522003.1
ENST00000523145.1
ENST00000414447.1
ENST00000377847.2
ENST00000377853.2
ENST00000377852.2
ENST00000523241.1
ENST00000520154.1
ENST00000358127.4
|
PAX5
|
paired box 5
|
|
chr14_+_50779029
|
0.068
|
ENST00000245448.6
|
ATP5S
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B)
|
|
chr2_-_70475730
|
0.067
|
ENST00000445587.1
ENST00000433529.2
ENST00000415783.2
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein
|
|
chr8_+_77593448
|
0.067
|
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr15_+_65822756
|
0.067
|
ENST00000562901.1
ENST00000261875.5
ENST00000442729.2
ENST00000565299.1
ENST00000568793.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1
|
|
chr2_+_45878790
|
0.066
|
ENST00000306156.3
|
PRKCE
|
protein kinase C, epsilon
|
|
chr12_-_6798523
|
0.066
|
ENST00000319770.3
|
ZNF384
|
zinc finger protein 384
|
|
chr5_+_56111361
|
0.064
|
ENST00000399503.3
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase
|
|
chr8_+_38614807
|
0.063
|
ENST00000330691.6
ENST00000348567.4
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr8_+_28351707
|
0.062
|
ENST00000537916.1
ENST00000523546.1
ENST00000240093.3
|
FZD3
|
frizzled family receptor 3
|
|
chr10_-_98346801
|
0.060
|
ENST00000371142.4
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr17_-_27278304
|
0.058
|
ENST00000577226.1
|
PHF12
|
PHD finger protein 12
|
|
chr3_-_72496035
|
0.058
|
ENST00000477973.2
|
RYBP
|
RING1 and YY1 binding protein
|
|
chr20_-_42939782
|
0.057
|
ENST00000396825.3
|
FITM2
|
fat storage-inducing transmembrane protein 2
|
|
chr1_+_36396677
|
0.057
|
ENST00000373191.4
ENST00000397828.2
|
AGO3
|
argonaute RISC catalytic component 3
|
|
chr1_-_39339777
|
0.054
|
ENST00000397572.2
|
MYCBP
|
MYC binding protein
|
|
chr12_-_14956396
|
0.054
|
ENST00000535328.1
ENST00000261167.2
|
WBP11
|
WW domain binding protein 11
|
|
chr14_-_22005343
|
0.054
|
ENST00000327430.3
|
SALL2
|
spalt-like transcription factor 2
|
|
chr15_+_40733387
|
0.052
|
ENST00000416165.1
|
BAHD1
|
bromo adjacent homology domain containing 1
|
|
chr10_+_97803151
|
0.052
|
ENST00000403870.3
ENST00000265992.5
ENST00000465148.2
ENST00000534974.1
|
CCNJ
|
cyclin J
|
|
chr15_-_35047166
|
0.049
|
ENST00000290374.4
|
GJD2
|
gap junction protein, delta 2, 36kDa
|
|
chr21_+_35445827
|
0.049
|
ENST00000381151.3
ENST00000608209.1
|
SLC5A3
SLC5A3
|
solute carrier family 5 (sodium/myo-inositol cotransporter), member 3
sodium/myo-inositol cotransporter
|
|
chr20_+_19193269
|
0.048
|
ENST00000328041.6
|
SLC24A3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3
|
|
chr8_-_71316021
|
0.045
|
ENST00000452400.2
|
NCOA2
|
nuclear receptor coactivator 2
|
|
chr19_-_38146289
|
0.045
|
ENST00000392144.1
ENST00000591444.1
ENST00000351218.2
ENST00000587809.1
|
ZFP30
|
ZFP30 zinc finger protein
|
|
chr20_-_34252759
|
0.044
|
ENST00000414711.1
ENST00000416778.1
ENST00000397442.1
ENST00000440240.1
ENST00000412056.1
ENST00000352393.4
ENST00000458038.1
ENST00000420363.1
ENST00000434795.1
ENST00000437100.1
ENST00000414664.1
ENST00000359646.1
ENST00000424458.1
ENST00000374104.3
ENST00000374114.3
|
CPNE1
RBM12
|
copine I
RNA binding motif protein 12
|
|
chr1_+_955448
|
0.043
|
ENST00000379370.2
|
AGRN
|
agrin
|
|
chr16_+_2587998
|
0.043
|
ENST00000441549.3
ENST00000268673.7
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr11_-_44331679
|
0.043
|
ENST00000329255.3
|
ALX4
|
ALX homeobox 4
|
|
chr12_+_121124599
|
0.043
|
ENST00000228506.3
|
MLEC
|
malectin
|
|
chrX_-_107979616
|
0.041
|
ENST00000372129.2
|
IRS4
|
insulin receptor substrate 4
|
|
chr1_-_155942086
|
0.040
|
ENST00000368315.4
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2
|
|
chr2_+_106361333
|
0.040
|
ENST00000233154.4
ENST00000451463.2
|
NCK2
|
NCK adaptor protein 2
|
|
chr2_+_27070964
|
0.039
|
ENST00000288699.6
|
DPYSL5
|
dihydropyrimidinase-like 5
|
|
chr20_+_56964169
|
0.039
|
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chr2_+_166095898
|
0.038
|
ENST00000424833.1
ENST00000375437.2
ENST00000357398.3
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr16_+_19078911
|
0.037
|
ENST00000321998.5
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast)
|
|
chr13_+_42622781
|
0.037
|
ENST00000337343.4
ENST00000261491.5
ENST00000379274.2
|
DGKH
|
diacylglycerol kinase, eta
|
|
chr6_-_30710510
|
0.036
|
ENST00000376389.3
|
FLOT1
|
flotillin 1
|
|
chr17_-_34890759
|
0.034
|
ENST00000431794.3
|
MYO19
|
myosin XIX
|
|
chr16_+_30710462
|
0.033
|
ENST00000262518.4
ENST00000395059.2
ENST00000344771.4
|
SRCAP
|
Snf2-related CREBBP activator protein
|
|
chr11_-_75062730
|
0.033
|
ENST00000420843.2
ENST00000360025.3
|
ARRB1
|
arrestin, beta 1
|
|
chr10_+_88516396
|
0.032
|
ENST00000372037.3
|
BMPR1A
|
bone morphogenetic protein receptor, type IA
|
|
chr10_-_13390270
|
0.030
|
ENST00000378614.4
ENST00000545675.1
ENST00000327347.5
|
SEPHS1
|
selenophosphate synthetase 1
|
|
chr3_+_14989076
|
0.029
|
ENST00000413118.1
ENST00000425241.1
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2
|
|
chrX_+_49020121
|
0.029
|
ENST00000415364.1
ENST00000376338.3
ENST00000425285.1
|
MAGIX
|
MAGI family member, X-linked
|
|
chr11_-_77532050
|
0.028
|
ENST00000308488.6
|
RSF1
|
remodeling and spacing factor 1
|
|
chr12_+_113229737
|
0.027
|
ENST00000551052.1
ENST00000415485.3
|
RPH3A
|
rabphilin 3A homolog (mouse)
|
|
chr1_+_199996702
|
0.027
|
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr11_+_73498898
|
0.026
|
ENST00000535529.1
ENST00000497094.2
ENST00000411840.2
ENST00000535277.1
ENST00000398483.3
ENST00000542303.1
|
MRPL48
|
mitochondrial ribosomal protein L48
|
|
chr6_-_17706618
|
0.025
|
ENST00000262077.2
ENST00000537253.1
|
NUP153
|
nucleoporin 153kDa
|
|
chr10_+_62538089
|
0.025
|
ENST00000519078.2
ENST00000395284.3
ENST00000316629.4
|
CDK1
|
cyclin-dependent kinase 1
|
|
chr17_+_4901199
|
0.025
|
ENST00000320785.5
ENST00000574165.1
|
KIF1C
|
kinesin family member 1C
|
|
chr6_+_163835669
|
0.024
|
ENST00000453779.2
ENST00000275262.7
ENST00000392127.2
ENST00000361752.3
|
QKI
|
QKI, KH domain containing, RNA binding
|
|
chr17_+_7184986
|
0.023
|
ENST00000317370.8
ENST00000571308.1
|
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4
|
|
chr3_+_14444063
|
0.023
|
ENST00000454876.2
ENST00000360861.3
ENST00000416216.2
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter), member 6
|
|
chr9_-_23821273
|
0.022
|
ENST00000380110.4
|
ELAVL2
|
ELAV like neuron-specific RNA binding protein 2
|
|
chr3_-_33686743
|
0.021
|
ENST00000333778.6
ENST00000539981.1
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr1_+_78245303
|
0.021
|
ENST00000370791.3
ENST00000443751.2
|
FAM73A
|
family with sequence similarity 73, member A
|
|
chrX_-_151619746
|
0.020
|
ENST00000370314.4
|
GABRA3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3
|
|
chr13_+_49280951
|
0.019
|
ENST00000282018.3
|
CYSLTR2
|
cysteinyl leukotriene receptor 2
|
|
chr15_+_79724858
|
0.019
|
ENST00000305428.3
|
KIAA1024
|
KIAA1024
|
|
chr11_-_6624801
|
0.018
|
ENST00000534343.1
ENST00000254605.6
|
RRP8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast)
|
|
chr20_+_42875887
|
0.018
|
ENST00000342560.5
|
GDAP1L1
|
ganglioside induced differentiation associated protein 1-like 1
|
|
chr6_-_116381918
|
0.017
|
ENST00000606080.1
|
FRK
|
fyn-related kinase
|
|
chr12_+_122516626
|
0.017
|
ENST00000319080.7
|
MLXIP
|
MLX interacting protein
|
|
chr2_-_218808771
|
0.016
|
ENST00000449814.1
ENST00000171887.4
|
TNS1
|
tensin 1
|
|
chr14_-_74551172
|
0.016
|
ENST00000553458.1
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr2_+_166326157
|
0.016
|
ENST00000421875.1
ENST00000314499.7
ENST00000409664.1
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr12_-_93835665
|
0.015
|
ENST00000552442.1
ENST00000550657.1
|
UBE2N
|
ubiquitin-conjugating enzyme E2N
|
|
chr2_-_178937478
|
0.015
|
ENST00000286063.6
|
PDE11A
|
phosphodiesterase 11A
|
|
chr14_-_75593708
|
0.015
|
ENST00000557673.1
ENST00000238616.5
|
NEK9
|
NIMA-related kinase 9
|
|
chr18_+_46065393
|
0.014
|
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor
|
|
chr2_-_153032484
|
0.014
|
ENST00000263904.4
|
STAM2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2
|
|
chr1_-_208417620
|
0.013
|
ENST00000367033.3
|
PLXNA2
|
plexin A2
|
|
chr22_+_31090793
|
0.012
|
ENST00000332585.6
ENST00000382310.3
ENST00000446658.2
|
OSBP2
|
oxysterol binding protein 2
|
|
chr16_+_69458428
|
0.012
|
ENST00000512062.1
ENST00000307892.8
|
CYB5B
|
cytochrome b5 type B (outer mitochondrial membrane)
|
|
chr12_+_113416191
|
0.012
|
ENST00000342315.4
ENST00000392583.2
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa
|
|
chr5_+_141303373
|
0.011
|
ENST00000432126.2
ENST00000194118.4
|
KIAA0141
|
KIAA0141
|
|
chr15_-_49447835
|
0.011
|
ENST00000388901.5
ENST00000299259.6
|
COPS2
|
COP9 signalosome subunit 2
|
|
chr1_+_89149905
|
0.011
|
ENST00000316005.7
ENST00000370521.3
ENST00000370505.3
|
PKN2
|
protein kinase N2
|
|
chr12_-_32908809
|
0.011
|
ENST00000324868.8
|
YARS2
|
tyrosyl-tRNA synthetase 2, mitochondrial
|
|
chr16_-_31021921
|
0.011
|
ENST00000215095.5
|
STX1B
|
syntaxin 1B
|
|
chr1_+_57110972
|
0.011
|
ENST00000371244.4
|
PRKAA2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit
|
|
chr5_-_137090028
|
0.010
|
ENST00000314940.4
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0
|
|
chr5_-_176778523
|
0.009
|
ENST00000513877.1
ENST00000515209.1
ENST00000514458.1
ENST00000502560.1
|
LMAN2
|
lectin, mannose-binding 2
|
|
chr16_-_57481278
|
0.008
|
ENST00000567751.1
ENST00000568940.1
ENST00000563341.1
ENST00000565961.1
ENST00000569370.1
ENST00000567518.1
ENST00000565786.1
ENST00000394391.4
|
CIAPIN1
|
cytokine induced apoptosis inhibitor 1
|
|
chr8_+_110346546
|
0.007
|
ENST00000521662.1
ENST00000521688.1
ENST00000520147.1
|
ENY2
|
enhancer of yellow 2 homolog (Drosophila)
|
|
chr14_+_75230011
|
0.007
|
ENST00000552421.1
ENST00000325680.7
ENST00000238571.3
|
YLPM1
|
YLP motif containing 1
|
|
chr2_-_38604398
|
0.006
|
ENST00000443098.1
ENST00000449130.1
ENST00000378954.4
ENST00000539122.1
ENST00000419554.2
ENST00000451483.1
ENST00000406122.1
|
ATL2
|
atlastin GTPase 2
|
|
chr1_-_24306798
|
0.006
|
ENST00000374452.5
ENST00000492112.2
ENST00000343255.5
ENST00000344989.6
|
SRSF10
|
serine/arginine-rich splicing factor 10
|
|
chr21_+_37858165
|
0.005
|
ENST00000595927.1
|
AP000695.1
|
AP000695.1
|
|
chr17_-_617949
|
0.004
|
ENST00000401468.3
ENST00000574029.1
ENST00000291074.5
ENST00000571805.1
ENST00000437048.2
ENST00000446250.2
|
VPS53
|
vacuolar protein sorting 53 homolog (S. cerevisiae)
|
|
chr9_+_115983808
|
0.004
|
ENST00000374210.6
ENST00000374212.4
|
SLC31A1
|
solute carrier family 31 (copper transporter), member 1
|
|
chr8_+_23104130
|
0.004
|
ENST00000313219.7
ENST00000519984.1
|
CHMP7
|
charged multivesicular body protein 7
|
|
chr12_+_56367697
|
0.004
|
ENST00000553116.1
ENST00000360299.5
ENST00000548068.1
ENST00000549915.1
ENST00000551459.1
ENST00000448789.2
|
RAB5B
|
RAB5B, member RAS oncogene family
|
|
chr16_+_70148230
|
0.004
|
ENST00000398122.3
ENST00000568530.1
|
PDPR
|
pyruvate dehydrogenase phosphatase regulatory subunit
|
|
chr1_-_39347255
|
0.003
|
ENST00000454994.2
ENST00000357771.3
|
GJA9
|
gap junction protein, alpha 9, 59kDa
|
|
chr16_+_4897632
|
0.002
|
ENST00000262376.6
|
UBN1
|
ubinuclein 1
|
|
chr6_+_28109703
|
0.002
|
ENST00000457389.2
ENST00000330236.6
|
ZKSCAN8
|
zinc finger with KRAB and SCAN domains 8
|
|
chr17_+_39845134
|
0.002
|
ENST00000591776.1
ENST00000469257.1
|
EIF1
|
eukaryotic translation initiation factor 1
|
|
chr2_+_204571198
|
0.002
|
ENST00000374481.3
ENST00000458610.2
ENST00000324106.8
|
CD28
|
CD28 molecule
|
|
chr7_+_92158083
|
0.001
|
ENST00000265732.5
ENST00000481551.1
ENST00000496410.1
|
RBM48
|
RNA binding motif protein 48
|
|
chr1_+_26560676
|
0.001
|
ENST00000451429.2
ENST00000252992.4
|
CEP85
|
centrosomal protein 85kDa
|
|
chr17_+_8213590
|
0.000
|
ENST00000361926.3
|
ARHGEF15
|
Rho guanine nucleotide exchange factor (GEF) 15
|