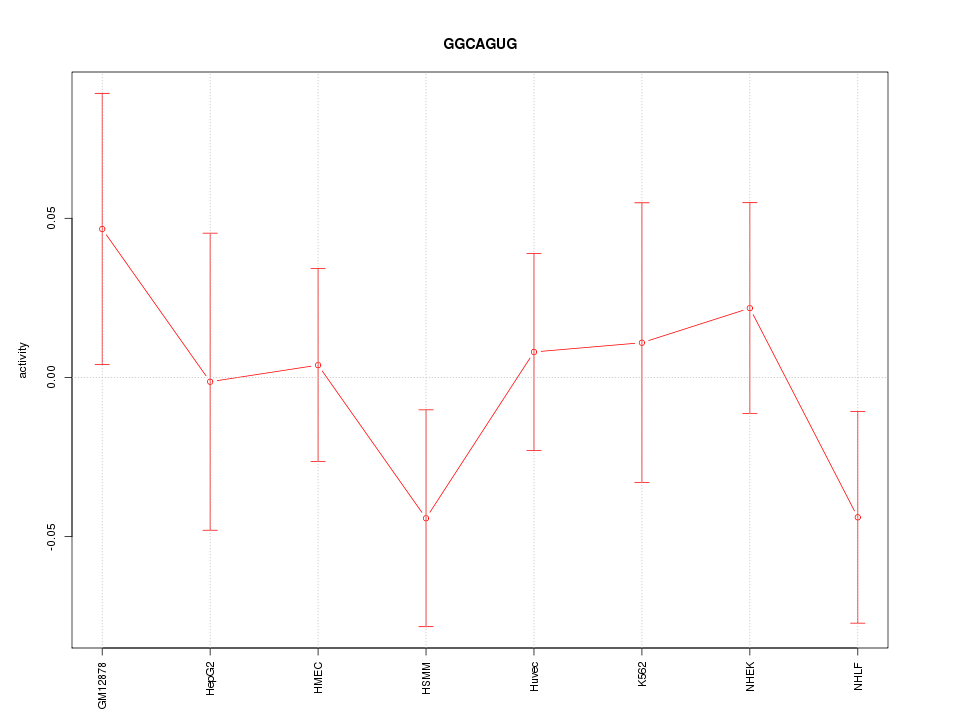
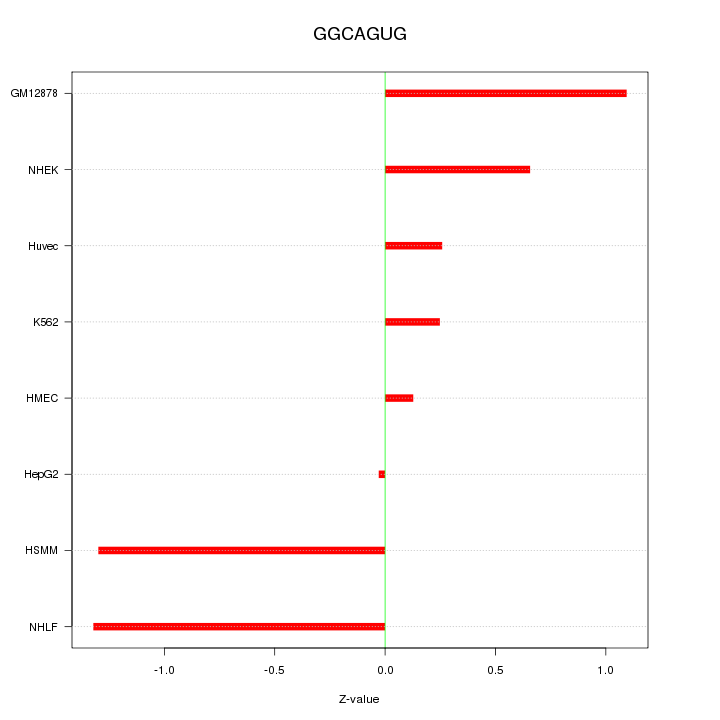
| 0.1 |
0.6 |
GO:0046671 |
regulation of nitrogen utilization(GO:0006808) positive regulation of neuron maturation(GO:0014042) nitrogen utilization(GO:0019740) cochlear nucleus development(GO:0021747) negative regulation of cellular pH reduction(GO:0032848) glial cell apoptotic process(GO:0034349) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) positive regulation of cell maturation(GO:1903431) |
| 0.1 |
0.4 |
GO:0003176 |
aortic valve development(GO:0003176) aortic valve morphogenesis(GO:0003180) positive regulation of endothelial cell differentiation(GO:0045603) positive regulation of keratinocyte differentiation(GO:0045618) regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 |
0.6 |
GO:0002328 |
pro-B cell differentiation(GO:0002328) |
| 0.1 |
0.4 |
GO:0048935 |
peripheral nervous system neuron development(GO:0048935) |
| 0.1 |
0.3 |
GO:0071539 |
Golgi localization(GO:0051645) protein localization to centrosome(GO:0071539) |
| 0.1 |
0.3 |
GO:0070544 |
histone H3-K36 demethylation(GO:0070544) |
| 0.1 |
0.2 |
GO:0003056 |
regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.1 |
1.0 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.1 |
0.2 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.1 |
1.0 |
GO:0071230 |
cellular response to amino acid stimulus(GO:0071230) |
| 0.1 |
0.2 |
GO:0061198 |
fungiform papilla development(GO:0061196) fungiform papilla morphogenesis(GO:0061197) fungiform papilla formation(GO:0061198) |
| 0.1 |
0.2 |
GO:0071294 |
cellular response to zinc ion(GO:0071294) |
| 0.1 |
0.2 |
GO:0060235 |
lens induction in camera-type eye(GO:0060235) |
| 0.1 |
0.3 |
GO:0018106 |
peptidyl-histidine phosphorylation(GO:0018106) |
| 0.1 |
0.2 |
GO:0018206 |
peptidyl-methionine modification(GO:0018206) |
| 0.1 |
0.2 |
GO:2000078 |
epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) regulation of type B pancreatic cell development(GO:2000074) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.1 |
0.2 |
GO:0055005 |
optic placode formation involved in camera-type eye formation(GO:0046619) ventricular cardiac myofibril assembly(GO:0055005) regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 |
0.1 |
GO:0030205 |
dermatan sulfate metabolic process(GO:0030205) dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 |
0.1 |
GO:0071109 |
superior temporal gyrus development(GO:0071109) |
| 0.0 |
0.1 |
GO:0051066 |
dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 |
0.2 |
GO:0071436 |
sodium ion export(GO:0071436) |
| 0.0 |
0.1 |
GO:0007028 |
cytoplasm organization(GO:0007028) |
| 0.0 |
0.1 |
GO:0032927 |
positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 |
0.3 |
GO:0021978 |
telencephalon regionalization(GO:0021978) |
| 0.0 |
0.2 |
GO:0032696 |
negative regulation of interleukin-13 production(GO:0032696) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 |
0.2 |
GO:0002430 |
complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 |
0.3 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.2 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 |
0.2 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.0 |
0.9 |
GO:0030851 |
granulocyte differentiation(GO:0030851) |
| 0.0 |
0.2 |
GO:0017055 |
negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.0 |
0.2 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
0.4 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.0 |
0.1 |
GO:0006668 |
sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 |
0.1 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 |
0.3 |
GO:0002089 |
lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 |
0.2 |
GO:0032075 |
positive regulation of nuclease activity(GO:0032075) |
| 0.0 |
0.1 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 |
0.1 |
GO:0071364 |
cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 |
0.2 |
GO:0060314 |
regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 |
0.2 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 |
0.2 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 |
0.4 |
GO:0006198 |
cAMP catabolic process(GO:0006198) |
| 0.0 |
0.1 |
GO:0033147 |
negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 |
0.2 |
GO:0006474 |
N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 |
0.1 |
GO:0009202 |
purine deoxyribonucleotide biosynthetic process(GO:0009153) deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.0 |
0.2 |
GO:0042354 |
fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 |
0.2 |
GO:0000183 |
chromatin silencing at rDNA(GO:0000183) |
| 0.0 |
0.1 |
GO:0035058 |
nonmotile primary cilium assembly(GO:0035058) |
| 0.0 |
0.2 |
GO:0060487 |
lung epithelial cell differentiation(GO:0060487) |
| 0.0 |
0.1 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) |
| 0.0 |
0.1 |
GO:0010586 |
miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 |
0.1 |
GO:0060676 |
ureteric bud formation(GO:0060676) mesonephric tubule formation(GO:0072172) |
| 0.0 |
0.2 |
GO:0051926 |
negative regulation of calcium ion transport(GO:0051926) |
| 0.0 |
0.2 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.0 |
0.2 |
GO:0006816 |
calcium ion transport(GO:0006816) |
| 0.0 |
0.2 |
GO:0001573 |
ganglioside metabolic process(GO:0001573) |
| 0.0 |
0.4 |
GO:0046847 |
filopodium assembly(GO:0046847) |
| 0.0 |
0.1 |
GO:0042816 |
pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 |
0.3 |
GO:0034644 |
cellular response to UV(GO:0034644) |
| 0.0 |
0.2 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 |
0.5 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.1 |
GO:0001554 |
luteolysis(GO:0001554) |
| 0.0 |
0.2 |
GO:0048384 |
retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 |
0.2 |
GO:0043097 |
pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 |
0.0 |
GO:0002043 |
blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 |
0.3 |
GO:0046834 |
lipid phosphorylation(GO:0046834) |
| 0.0 |
0.7 |
GO:0030183 |
B cell differentiation(GO:0030183) |
| 0.0 |
0.0 |
GO:0046579 |
positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 |
0.1 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |