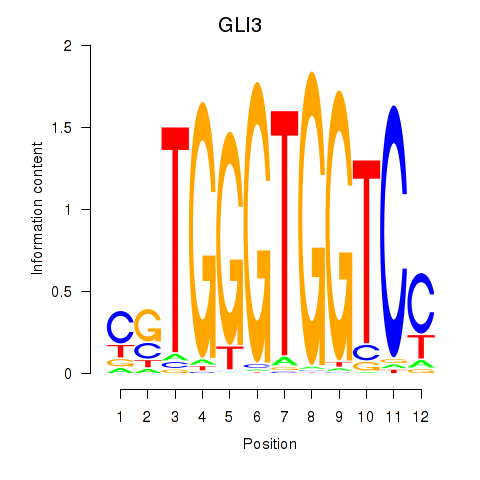
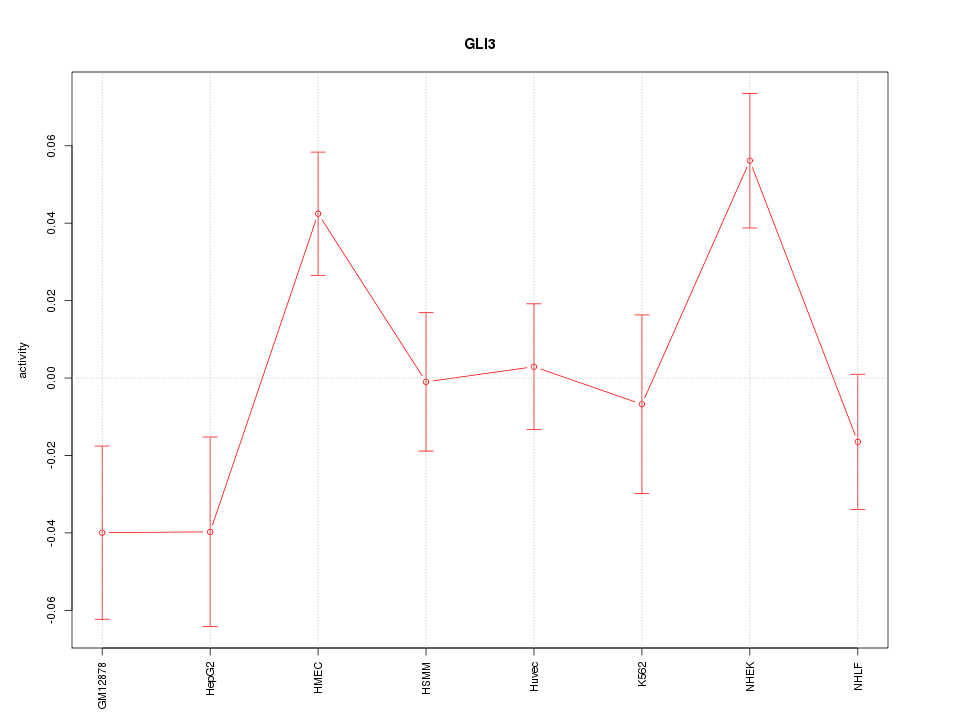
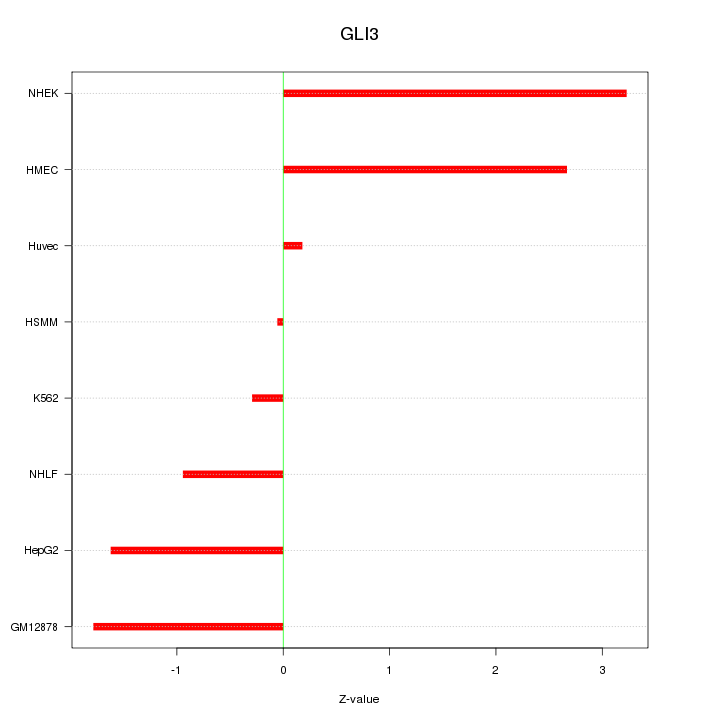
| 1.2 |
3.7 |
GO:0071603 |
negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) endothelial cell-cell adhesion(GO:0071603) |
| 0.9 |
2.7 |
GO:0010701 |
positive regulation of chronic inflammatory response(GO:0002678) positive regulation of norepinephrine secretion(GO:0010701) |
| 0.6 |
1.7 |
GO:2000064 |
regulation of cortisol biosynthetic process(GO:2000064) |
| 0.6 |
2.2 |
GO:0043128 |
regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.5 |
1.6 |
GO:0019367 |
fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.5 |
14.3 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.5 |
2.3 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.4 |
2.2 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.4 |
1.2 |
GO:0019747 |
regulation of isoprenoid metabolic process(GO:0019747) |
| 0.3 |
1.4 |
GO:0042335 |
cuticle development(GO:0042335) |
| 0.3 |
8.2 |
GO:0045745 |
positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.3 |
1.0 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.3 |
2.0 |
GO:0002052 |
positive regulation of neuroblast proliferation(GO:0002052) |
| 0.2 |
1.0 |
GO:0030913 |
paranodal junction assembly(GO:0030913) |
| 0.2 |
0.7 |
GO:0060279 |
progesterone secretion(GO:0042701) regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.2 |
0.2 |
GO:0051589 |
negative regulation of neurotransmitter transport(GO:0051589) |
| 0.2 |
0.7 |
GO:0009202 |
purine deoxyribonucleotide biosynthetic process(GO:0009153) deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.2 |
0.9 |
GO:0060662 |
tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.2 |
6.3 |
GO:0007398 |
ectoderm development(GO:0007398) |
| 0.2 |
0.9 |
GO:0060708 |
spongiotrophoblast differentiation(GO:0060708) |
| 0.2 |
0.6 |
GO:0043371 |
negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T-helper cell differentiation(GO:0045623) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.2 |
0.6 |
GO:0006788 |
heme oxidation(GO:0006788) |
| 0.2 |
1.8 |
GO:0051552 |
flavone metabolic process(GO:0051552) |
| 0.2 |
1.9 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.2 |
3.0 |
GO:0050812 |
acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.2 |
0.2 |
GO:0050653 |
chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.2 |
2.0 |
GO:0010658 |
striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.2 |
0.5 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 |
0.5 |
GO:0010248 |
establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) post-embryonic camera-type eye development(GO:0031077) |
| 0.1 |
0.9 |
GO:2000124 |
cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 |
0.9 |
GO:0005984 |
disaccharide metabolic process(GO:0005984) polysaccharide digestion(GO:0044245) |
| 0.1 |
0.6 |
GO:0032926 |
negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 |
0.8 |
GO:0060325 |
luteinization(GO:0001553) face morphogenesis(GO:0060325) |
| 0.1 |
0.5 |
GO:0048295 |
positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.1 |
1.5 |
GO:0060438 |
trachea development(GO:0060438) |
| 0.1 |
0.7 |
GO:0090025 |
regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.1 |
0.5 |
GO:0046022 |
regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.1 |
0.4 |
GO:0043461 |
proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 |
1.1 |
GO:0036336 |
dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.1 |
1.5 |
GO:0008631 |
intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) cell death in response to oxidative stress(GO:0036473) |
| 0.1 |
0.3 |
GO:0032938 |
negative regulation of translation in response to oxidative stress(GO:0032938) regulation of translation in response to oxidative stress(GO:0043556) |
| 0.1 |
0.5 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.1 |
0.8 |
GO:0035313 |
wound healing, spreading of epidermal cells(GO:0035313) wound healing, spreading of cells(GO:0044319) epiboly(GO:0090504) epiboly involved in wound healing(GO:0090505) |
| 0.1 |
1.6 |
GO:0051016 |
barbed-end actin filament capping(GO:0051016) |
| 0.1 |
3.2 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.1 |
0.5 |
GO:0002220 |
innate immune response activating cell surface receptor signaling pathway(GO:0002220) |
| 0.1 |
0.6 |
GO:0045071 |
negative regulation of viral genome replication(GO:0045071) |
| 0.1 |
1.1 |
GO:0010799 |
regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 |
1.0 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 |
0.4 |
GO:0034204 |
lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 |
0.4 |
GO:0016199 |
axon midline choice point recognition(GO:0016199) |
| 0.1 |
0.5 |
GO:0044413 |
evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 |
0.5 |
GO:0006295 |
nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 |
0.6 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 |
0.5 |
GO:0006658 |
phosphatidylserine metabolic process(GO:0006658) |
| 0.1 |
0.6 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.1 |
0.3 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.1 |
0.2 |
GO:0090598 |
male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.1 |
0.7 |
GO:0001946 |
lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 0.1 |
0.1 |
GO:0060298 |
positive regulation of sarcomere organization(GO:0060298) |
| 0.1 |
0.1 |
GO:0033632 |
regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 |
5.8 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |
| 0.1 |
0.2 |
GO:0051547 |
regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.1 |
0.1 |
GO:0030202 |
heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 |
0.4 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 |
0.3 |
GO:0007341 |
penetration of zona pellucida(GO:0007341) |
| 0.1 |
0.7 |
GO:0006702 |
androgen biosynthetic process(GO:0006702) |
| 0.1 |
5.8 |
GO:0043542 |
endothelial cell migration(GO:0043542) |
| 0.1 |
0.3 |
GO:0014820 |
tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.1 |
2.3 |
GO:0006309 |
DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.1 |
1.4 |
GO:0042730 |
fibrinolysis(GO:0042730) |
| 0.1 |
0.2 |
GO:1902803 |
regulation of synaptic vesicle transport(GO:1902803) regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 |
1.1 |
GO:0007405 |
neuroblast proliferation(GO:0007405) |
| 0.1 |
0.3 |
GO:0070682 |
proteasome regulatory particle assembly(GO:0070682) |
| 0.1 |
0.2 |
GO:0033602 |
negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of dopamine secretion(GO:0033602) |
| 0.1 |
0.5 |
GO:0060716 |
labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 |
1.2 |
GO:0007032 |
endosome organization(GO:0007032) |
| 0.1 |
0.2 |
GO:1903960 |
negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.1 |
0.1 |
GO:0002295 |
T-helper cell lineage commitment(GO:0002295) positive regulation of cell fate commitment(GO:0010455) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) regulation of T-helper 17 type immune response(GO:2000316) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 17 cell differentiation(GO:2000319) positive regulation of T-helper 17 cell differentiation(GO:2000321) regulation of T-helper 17 cell lineage commitment(GO:2000328) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.1 |
0.5 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 |
0.7 |
GO:0051927 |
obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.1 |
1.8 |
GO:0008633 |
obsolete activation of pro-apoptotic gene products(GO:0008633) |
| 0.1 |
0.9 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.1 |
0.3 |
GO:0002430 |
complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 |
0.2 |
GO:0006530 |
asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.1 |
3.7 |
GO:0034332 |
adherens junction organization(GO:0034332) |
| 0.0 |
0.2 |
GO:0002860 |
natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 |
0.6 |
GO:0090398 |
cellular senescence(GO:0090398) |
| 0.0 |
0.1 |
GO:0051102 |
DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 |
0.4 |
GO:0046835 |
carbohydrate phosphorylation(GO:0046835) |
| 0.0 |
0.3 |
GO:0030007 |
cellular potassium ion homeostasis(GO:0030007) negative regulation of cell volume(GO:0045794) |
| 0.0 |
0.5 |
GO:0010633 |
negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 |
0.3 |
GO:0097384 |
cellular lipid biosynthetic process(GO:0097384) |
| 0.0 |
0.2 |
GO:1903008 |
organelle disassembly(GO:1903008) |
| 0.0 |
0.2 |
GO:0071025 |
RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 |
0.3 |
GO:0046349 |
UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.0 |
0.3 |
GO:0006561 |
proline biosynthetic process(GO:0006561) |
| 0.0 |
0.1 |
GO:0032621 |
interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.0 |
0.5 |
GO:1904031 |
positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 |
0.2 |
GO:0045948 |
positive regulation of translational initiation(GO:0045948) |
| 0.0 |
0.2 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.0 |
0.1 |
GO:0035385 |
Roundabout signaling pathway(GO:0035385) response to cortisol(GO:0051414) apoptotic process involved in luteolysis(GO:0061364) regulation of chemokine-mediated signaling pathway(GO:0070099) negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 |
0.2 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.0 |
2.6 |
GO:0006939 |
smooth muscle contraction(GO:0006939) |
| 0.0 |
0.5 |
GO:0050872 |
white fat cell differentiation(GO:0050872) |
| 0.0 |
0.2 |
GO:0046676 |
negative regulation of insulin secretion(GO:0046676) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.0 |
0.3 |
GO:0000266 |
mitochondrial fission(GO:0000266) |
| 0.0 |
0.1 |
GO:0050747 |
positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 |
0.5 |
GO:0006449 |
regulation of translational termination(GO:0006449) |
| 0.0 |
0.0 |
GO:0042044 |
water transport(GO:0006833) fluid transport(GO:0042044) |
| 0.0 |
0.2 |
GO:0002063 |
chondrocyte development(GO:0002063) |
| 0.0 |
0.1 |
GO:0042268 |
regulation of cytolysis(GO:0042268) |
| 0.0 |
0.2 |
GO:0050689 |
negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 |
2.1 |
GO:0006821 |
chloride transport(GO:0006821) |
| 0.0 |
0.1 |
GO:0042986 |
positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 |
0.2 |
GO:0032472 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) Golgi calcium ion transport(GO:0032472) manganese ion homeostasis(GO:0055071) |
| 0.0 |
0.1 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.0 |
0.1 |
GO:0051958 |
methotrexate transport(GO:0051958) |
| 0.0 |
0.3 |
GO:0000183 |
chromatin silencing at rDNA(GO:0000183) |
| 0.0 |
0.7 |
GO:0045103 |
intermediate filament-based process(GO:0045103) |
| 0.0 |
0.7 |
GO:0008299 |
isoprenoid biosynthetic process(GO:0008299) |
| 0.0 |
0.4 |
GO:0031000 |
response to caffeine(GO:0031000) |
| 0.0 |
0.4 |
GO:0008088 |
axo-dendritic transport(GO:0008088) |
| 0.0 |
0.1 |
GO:0072385 |
minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 |
0.1 |
GO:0006423 |
cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 |
0.1 |
GO:1902414 |
protein localization to adherens junction(GO:0071896) protein localization to cell junction(GO:1902414) |
| 0.0 |
0.1 |
GO:0018352 |
glutamate decarboxylation to succinate(GO:0006540) protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 |
0.1 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 |
0.3 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
0.2 |
GO:0048630 |
skeletal muscle tissue growth(GO:0048630) |
| 0.0 |
0.2 |
GO:0060479 |
lung epithelium development(GO:0060428) lung cell differentiation(GO:0060479) |
| 0.0 |
0.6 |
GO:0043154 |
negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.0 |
0.2 |
GO:0048892 |
sensory system development(GO:0048880) lateral line nerve development(GO:0048892) lateral line nerve glial cell differentiation(GO:0048895) lateral line system development(GO:0048925) lateral line nerve glial cell development(GO:0048937) iridophore differentiation(GO:0050935) |
| 0.0 |
0.2 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.0 |
0.0 |
GO:0002385 |
innate immune response in mucosa(GO:0002227) mucosal immune response(GO:0002385) |
| 0.0 |
0.1 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.0 |
0.3 |
GO:0008272 |
sulfate transport(GO:0008272) |
| 0.0 |
0.1 |
GO:0015705 |
iodide transport(GO:0015705) |
| 0.0 |
1.2 |
GO:0045839 |
negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 |
0.1 |
GO:0030186 |
melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 |
1.2 |
GO:0000079 |
regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 |
0.3 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 |
0.1 |
GO:0090201 |
negative regulation of mitochondrion organization(GO:0010823) response to cycloheximide(GO:0046898) negative regulation of release of cytochrome c from mitochondria(GO:0090201) negative regulation of apoptotic signaling pathway(GO:2001234) |
| 0.0 |
0.1 |
GO:0048537 |
mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 |
0.1 |
GO:0015670 |
carbon dioxide transport(GO:0015670) |
| 0.0 |
0.7 |
GO:0033572 |
ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 |
0.0 |
GO:0003077 |
obsolete negative regulation of diuresis(GO:0003077) |
| 0.0 |
0.3 |
GO:0014003 |
oligodendrocyte development(GO:0014003) |
| 0.0 |
0.3 |
GO:0018298 |
protein-chromophore linkage(GO:0018298) |
| 0.0 |
0.1 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 |
0.3 |
GO:0060393 |
regulation of pathway-restricted SMAD protein phosphorylation(GO:0060393) |
| 0.0 |
0.1 |
GO:0021516 |
dorsal spinal cord development(GO:0021516) |
| 0.0 |
0.2 |
GO:0043124 |
negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 |
0.3 |
GO:0003407 |
neural retina development(GO:0003407) |
| 0.0 |
0.4 |
GO:0016254 |
preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 |
0.1 |
GO:0034260 |
negative regulation of GTPase activity(GO:0034260) |
| 0.0 |
0.1 |
GO:0045008 |
depyrimidination(GO:0045008) |
| 0.0 |
0.1 |
GO:0006707 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.2 |
GO:0007494 |
midgut development(GO:0007494) |
| 0.0 |
0.4 |
GO:0015813 |
L-glutamate transport(GO:0015813) |
| 0.0 |
0.2 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.0 |
0.3 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.4 |
GO:0070830 |
bicellular tight junction assembly(GO:0070830) |
| 0.0 |
0.3 |
GO:0000305 |
response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) |
| 0.0 |
0.1 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.1 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 |
1.1 |
GO:0007605 |
sensory perception of sound(GO:0007605) |
| 0.0 |
0.4 |
GO:0042147 |
retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 |
0.1 |
GO:0044254 |
angiotensin catabolic process in blood(GO:0002005) hormone catabolic process(GO:0042447) multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 |
0.2 |
GO:0000045 |
autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 |
0.1 |
GO:0009396 |
folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 |
0.1 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
0.1 |
GO:0043306 |
positive regulation of mast cell activation(GO:0033005) positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 |
0.0 |
GO:0032506 |
barrier septum assembly(GO:0000917) cytokinetic process(GO:0032506) cell septum assembly(GO:0090529) mitotic cytokinetic process(GO:1902410) |
| 0.0 |
0.1 |
GO:0032096 |
negative regulation of response to food(GO:0032096) |
| 0.0 |
0.1 |
GO:0050435 |
beta-amyloid metabolic process(GO:0050435) |
| 0.0 |
0.4 |
GO:0001667 |
ameboidal-type cell migration(GO:0001667) |
| 0.0 |
0.6 |
GO:0043484 |
regulation of RNA splicing(GO:0043484) |
| 0.0 |
0.1 |
GO:0006338 |
chromatin remodeling(GO:0006338) |
| 0.0 |
0.2 |
GO:0007220 |
Notch receptor processing(GO:0007220) membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 |
0.1 |
GO:0032922 |
circadian regulation of gene expression(GO:0032922) |
| 0.0 |
0.1 |
GO:0032516 |
positive regulation of phosphoprotein phosphatase activity(GO:0032516) |