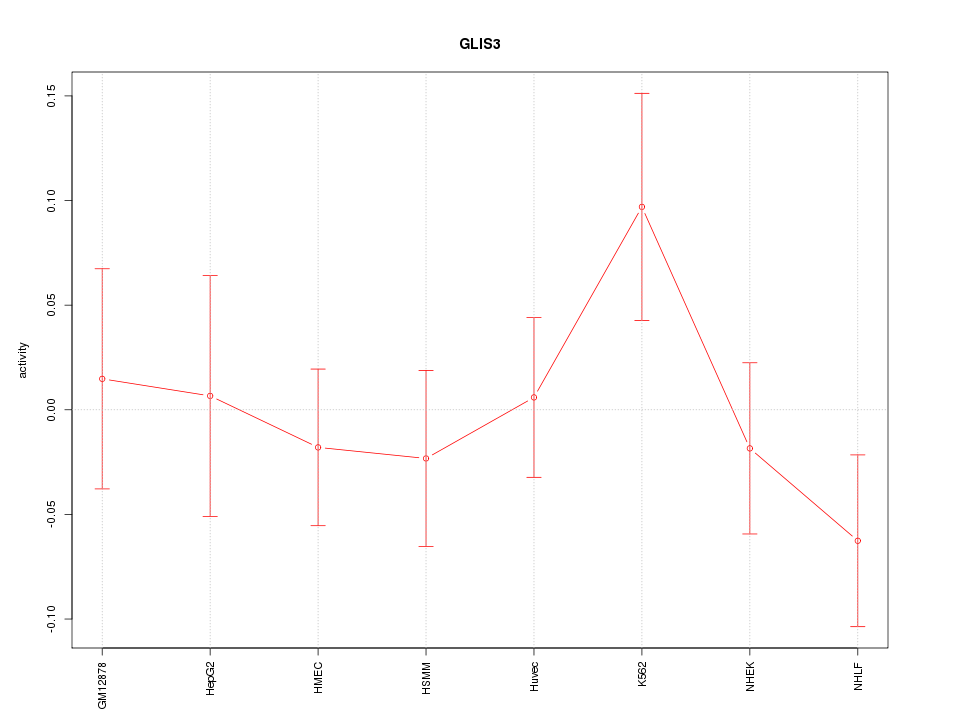
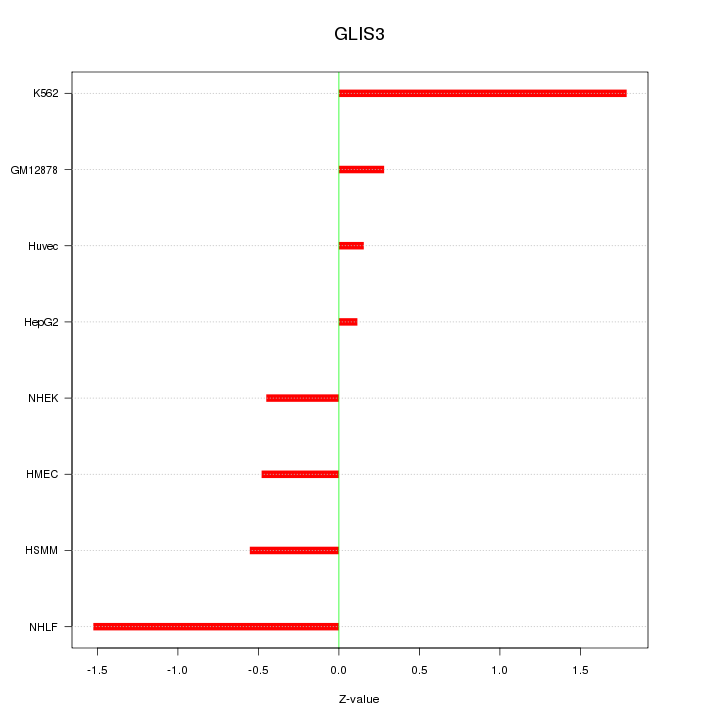
Motif ID: GLIS3
Z-value: 0.893
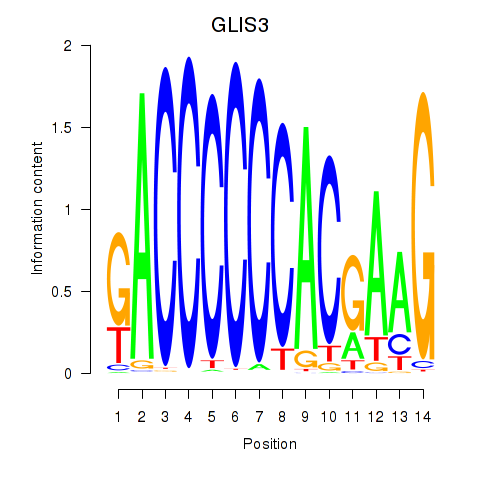
Transcription factors associated with GLIS3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| GLIS3 | ENSG00000107249.17 | GLIS3 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.3 | 0.8 | GO:0046732 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by virus of host immune response(GO:0075528) connective tissue replacement(GO:0097709) |
| 0.2 | 2.0 | GO:0031943 | regulation of glucocorticoid metabolic process(GO:0031943) |
| 0.2 | 0.5 | GO:0060318 | definitive erythrocyte differentiation(GO:0060318) |
| 0.2 | 0.5 | GO:0010586 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.1 | 1.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 1.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.4 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.1 | 0.3 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 1.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.8 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.2 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.3 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.2 | GO:0061756 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.8 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.0 | GO:0009279 | cell outer membrane(GO:0009279) |
| 0.0 | 0.8 | GO:0031901 | early endosome membrane(GO:0031901) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.5 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 1.1 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 2.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 2.6 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.1 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.3 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.5 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 1.0 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 1.3 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.0 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.4 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |