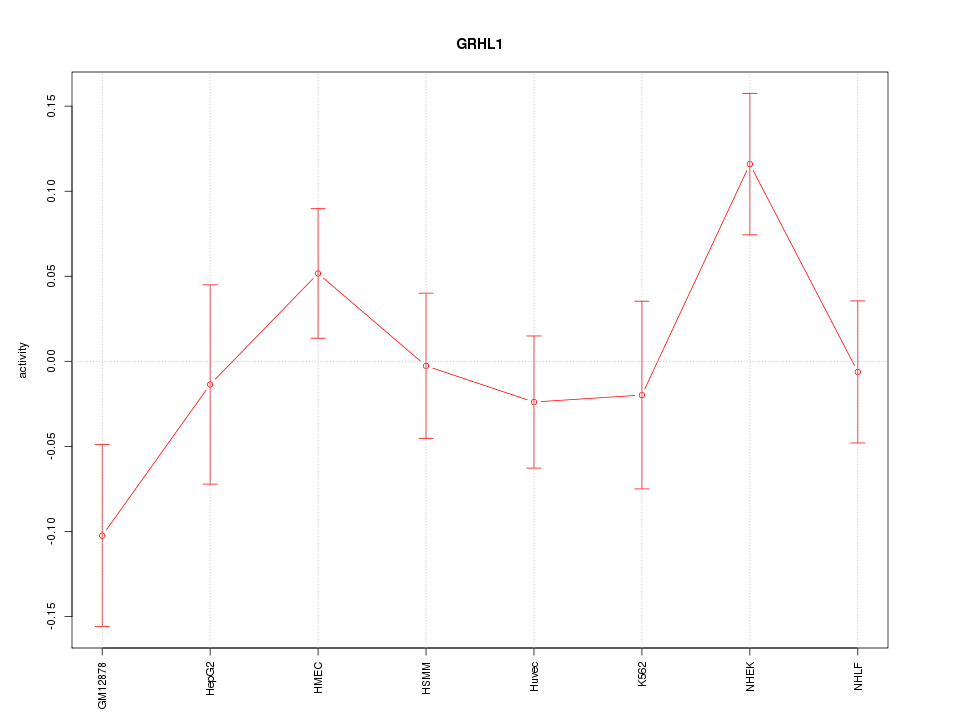
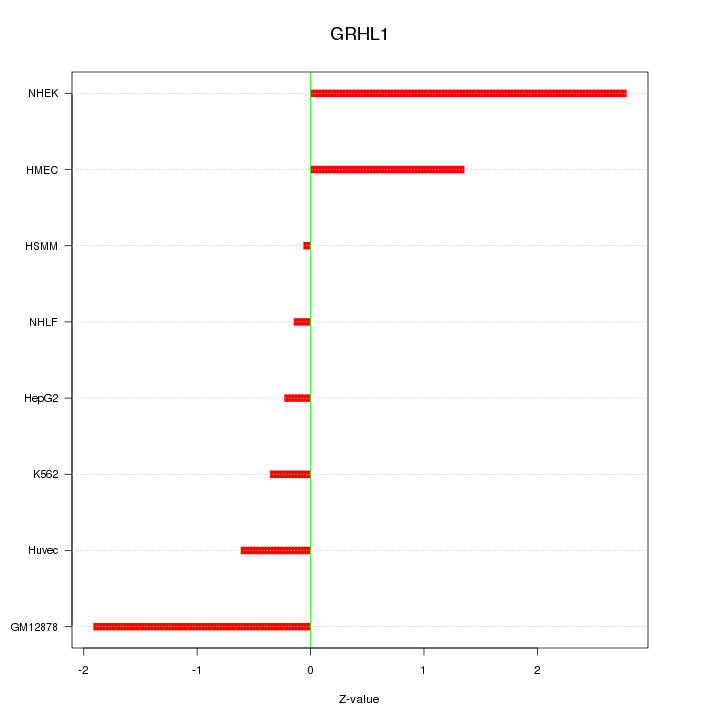
Motif ID: GRHL1
Z-value: 1.317
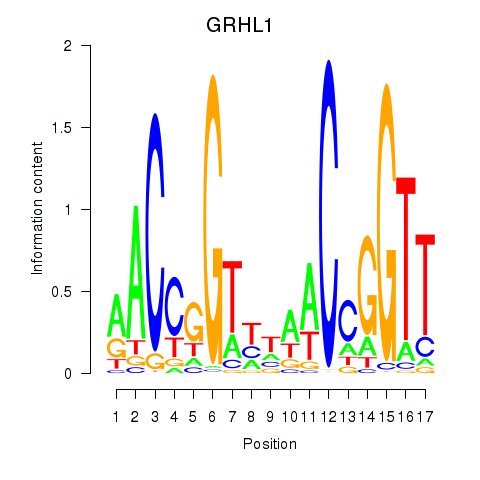
Transcription factors associated with GRHL1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| GRHL1 | ENSG00000134317.13 | GRHL1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.4 | 1.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.2 | 0.6 | GO:0006740 | glyoxylate cycle(GO:0006097) NADPH regeneration(GO:0006740) |
| 0.1 | 1.2 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.3 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 1.1 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 3.7 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:0060317 | cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.0 | 0.1 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.8 | GO:0033572 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 | 0.2 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 1.3 | GO:0006665 | sphingolipid metabolic process(GO:0006665) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.8 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 1.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.7 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.5 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 3.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0019861 | obsolete flagellum(GO:0019861) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 3.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 0.6 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.2 | 1.3 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 2.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.5 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) snoRNA binding(GO:0030515) |