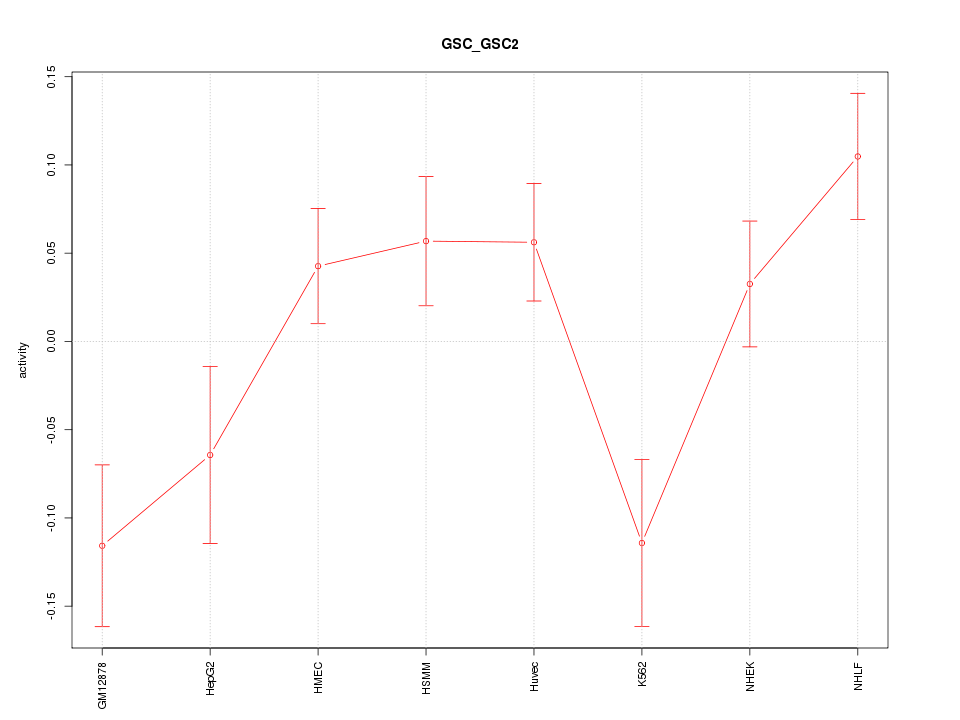
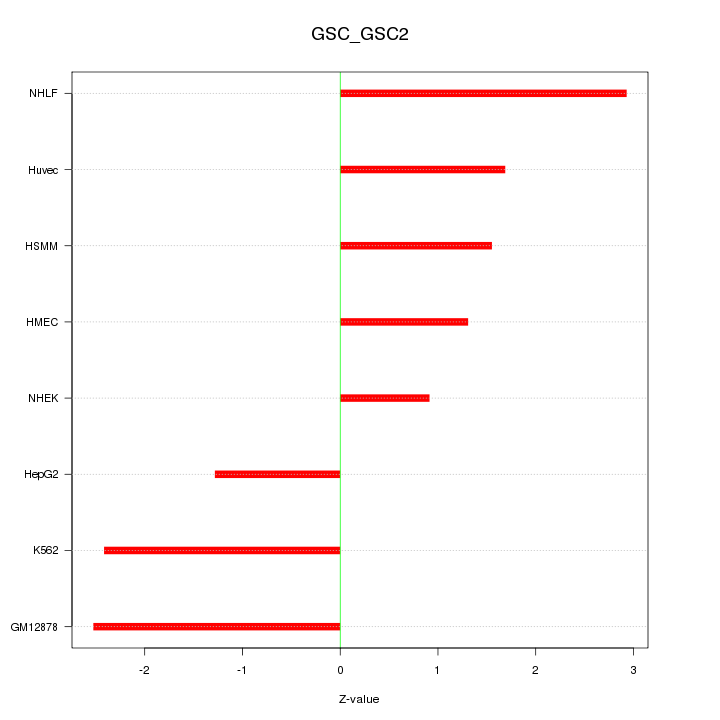
Motif ID: GSC_GSC2
Z-value: 1.945
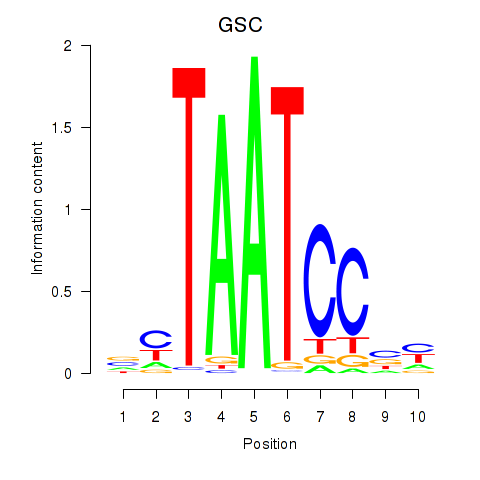
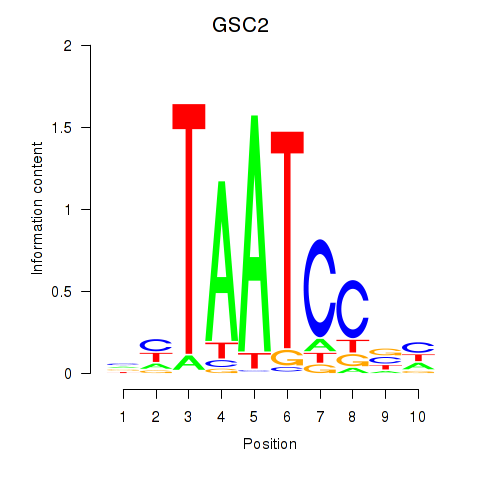
Transcription factors associated with GSC_GSC2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| GSC | ENSG00000133937.3 | GSC |
| GSC2 | ENSG00000063515.2 | GSC2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 21.3 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.7 | 3.0 | GO:0051503 | purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) |
| 0.3 | 3.0 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.3 | 2.6 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.2 | 1.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.2 | 0.7 | GO:0002124 | territorial aggressive behavior(GO:0002124) brainstem development(GO:0003360) vocalization behavior(GO:0071625) |
| 0.2 | 0.7 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 8.9 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 0.9 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.2 | 1.6 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.2 | 0.7 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.2 | 0.3 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 | 3.2 | GO:0050927 | positive regulation of positive chemotaxis(GO:0050927) |
| 0.1 | 2.4 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.1 | 3.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 2.0 | GO:0006911 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.1 | 2.2 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.1 | 0.4 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 0.6 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.4 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 4.4 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 2.2 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.0 | 0.6 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 7.7 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.3 | GO:0033523 | histone monoubiquitination(GO:0010390) histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.7 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.6 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.4 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 1.0 | GO:0051297 | centrosome organization(GO:0051297) |
| 0.0 | 0.5 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 1.0 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 5.1 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.3 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 1.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.5 | GO:0016101 | retinoid metabolic process(GO:0001523) diterpenoid metabolic process(GO:0016101) |
| 0.0 | 0.0 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 20.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.3 | 3.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 8.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 5.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 1.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.0 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 3.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 4.7 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 3.0 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.8 | 2.4 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.5 | 3.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 21.3 | GO:0051287 | NAD binding(GO:0051287) |
| 0.2 | 3.0 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 0.7 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.2 | 3.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 4.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.6 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.1 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.7 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 1.5 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 0.4 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 7.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 8.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.1 | 0.9 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.1 | 0.6 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.3 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.0 | 1.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.3 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 2.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0043022 | ribosome binding(GO:0043022) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.5 | SA_FAS_SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |