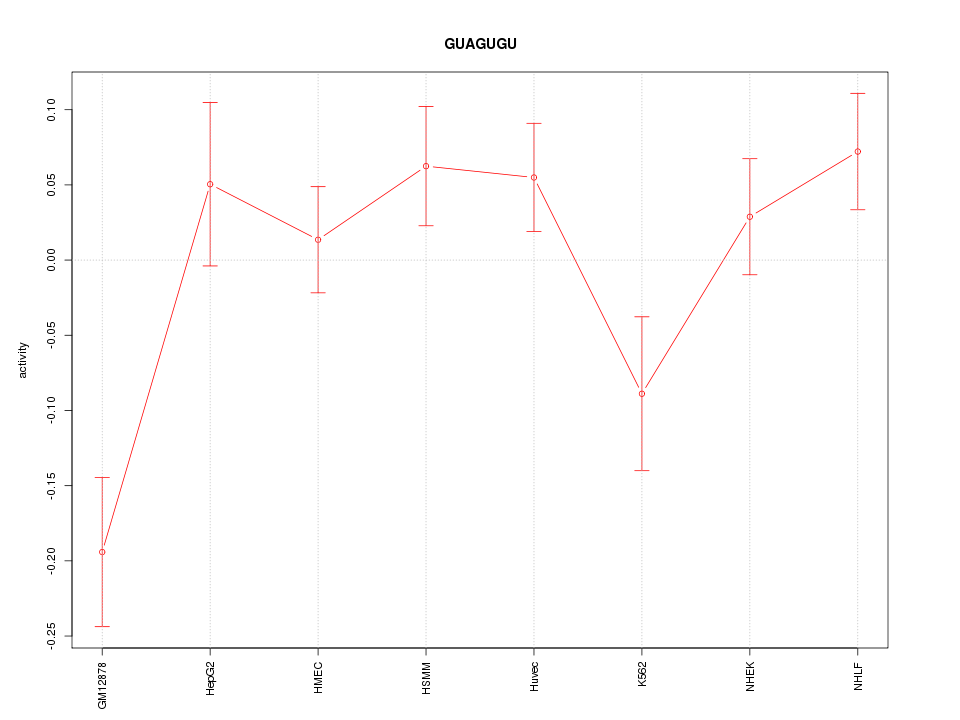
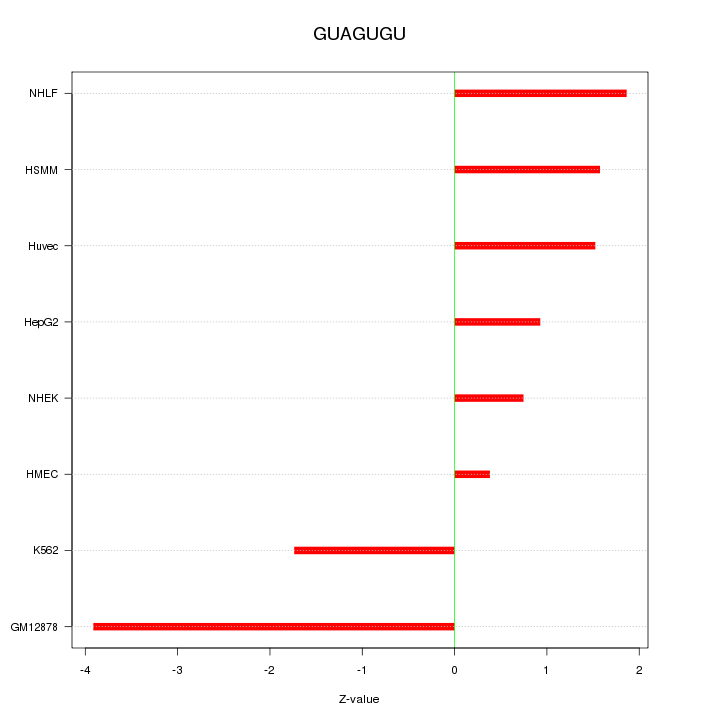
Motif ID: GUAGUGU
Z-value: 1.877

Mature miRNA associated with seed GUAGUGU:
| Name | miRBase Accession |
|---|---|
| hsa-miR-142-3p.1 | MIMAT0000434 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.6 | 4.4 | GO:0060414 | aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.4 | 1.3 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.3 | 0.8 | GO:0070672 | response to interleukin-12(GO:0070671) response to interleukin-15(GO:0070672) |
| 0.3 | 1.4 | GO:0046881 | positive regulation of gonadotropin secretion(GO:0032278) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.3 | 3.5 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.3 | 1.3 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.3 | 0.8 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.3 | 0.8 | GO:1902745 | positive regulation of lamellipodium assembly(GO:0010592) negative regulation of interleukin-23 production(GO:0032707) positive regulation of lamellipodium organization(GO:1902745) |
| 0.2 | 0.7 | GO:0043276 | anoikis(GO:0043276) |
| 0.2 | 0.8 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.2 | 0.6 | GO:0007028 | cytoplasm organization(GO:0007028) ribonucleoprotein complex disassembly(GO:0032988) |
| 0.2 | 0.7 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 1.5 | GO:0016322 | neuron remodeling(GO:0016322) regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.1 | 1.9 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 0.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 0.6 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.1 | 0.3 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.1 | 1.8 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 1.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 | 0.3 | GO:0045726 | negative regulation of macrophage cytokine production(GO:0010936) regulation of integrin biosynthetic process(GO:0045113) positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 2.1 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.1 | 0.2 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.1 | 0.4 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 | 3.0 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.1 | 0.4 | GO:0070498 | response to peptidoglycan(GO:0032494) interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 1.3 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.7 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.5 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.4 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.9 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.0 | 0.1 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 5.1 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 2.4 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.4 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 1.0 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.1 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) camera-type eye photoreceptor cell differentiation(GO:0060219) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.6 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.3 | 0.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 0.8 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 0.7 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.5 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 0.7 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.4 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 1.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 2.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 0.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.4 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.9 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 1.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.2 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 2.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 2.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.3 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 4.1 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 1.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.9 | GO:0005925 | focal adhesion(GO:0005925) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.7 | 3.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.3 | 1.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.3 | 0.8 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.2 | 0.7 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.2 | 2.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 0.6 | GO:0048184 | obsolete follistatin binding(GO:0048184) |
| 0.1 | 0.4 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.6 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.7 | GO:0052658 | PH domain binding(GO:0042731) inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.1 | 2.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.8 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.1 | 0.8 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.4 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 1.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 1.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 3.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.7 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 1.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.3 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.2 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.4 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 0.1 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.3 | GO:0008135 | translation factor activity, RNA binding(GO:0008135) |
| 0.0 | 1.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.9 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.4 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.0 | SIG_BCR_SIGNALING_PATHWAY | Members of the BCR signaling pathway |