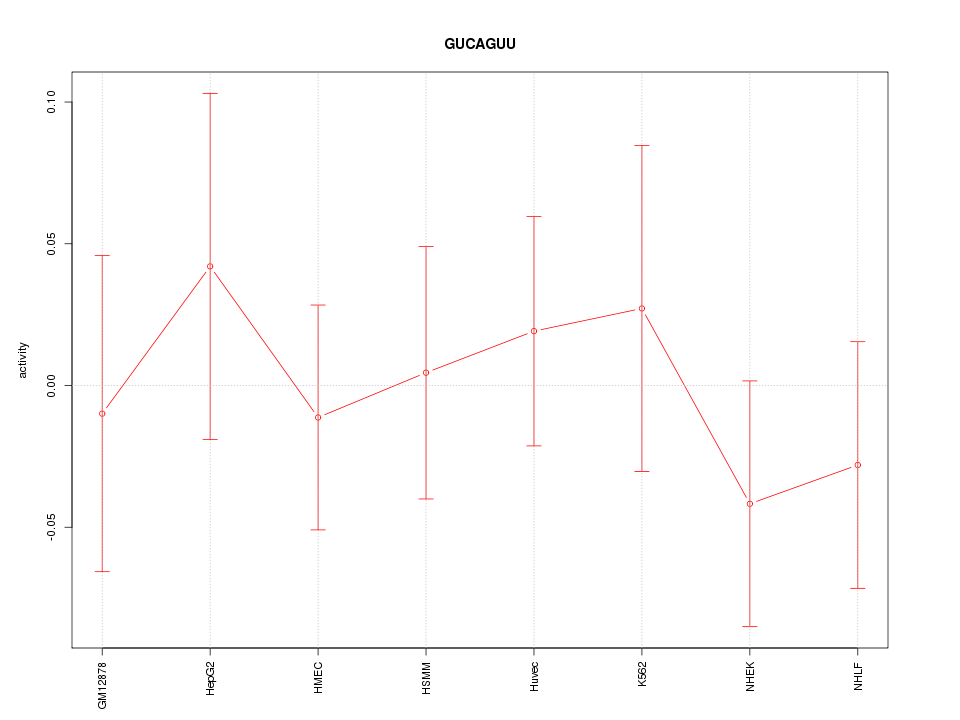
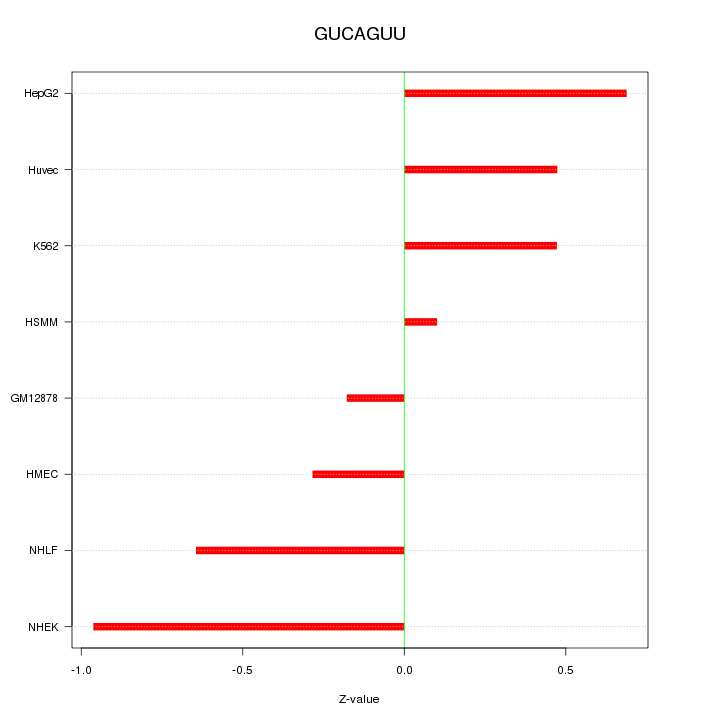
|
chr17_+_53342311
|
0.857
|
ENST00000226067.5
|
HLF
|
hepatic leukemia factor
|
|
chr11_-_86666427
|
0.426
|
ENST00000531380.1
|
FZD4
|
frizzled family receptor 4
|
|
chr1_-_92351769
|
0.405
|
ENST00000212355.4
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr13_-_76056250
|
0.326
|
ENST00000377636.3
ENST00000431480.2
ENST00000377625.2
ENST00000425511.1
|
TBC1D4
|
TBC1 domain family, member 4
|
|
chr1_+_93913713
|
0.315
|
ENST00000604705.1
ENST00000370253.2
|
FNBP1L
|
formin binding protein 1-like
|
|
chr8_+_81397876
|
0.303
|
ENST00000430430.1
|
ZBTB10
|
zinc finger and BTB domain containing 10
|
|
chr16_+_22308717
|
0.302
|
ENST00000299853.5
ENST00000564209.1
ENST00000565358.1
ENST00000418581.2
ENST00000564883.1
ENST00000359210.4
ENST00000563024.1
|
POLR3E
|
polymerase (RNA) III (DNA directed) polypeptide E (80kD)
|
|
chr15_-_68498376
|
0.291
|
ENST00000540479.1
ENST00000395465.3
|
CALML4
|
calmodulin-like 4
|
|
chr5_-_126366500
|
0.279
|
ENST00000308660.5
|
MARCH3
|
membrane-associated ring finger (C3HC4) 3, E3 ubiquitin protein ligase
|
|
chr6_-_79787902
|
0.265
|
ENST00000275034.4
|
PHIP
|
pleckstrin homology domain interacting protein
|
|
chr11_-_33891362
|
0.261
|
ENST00000395833.3
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr1_-_182361327
|
0.241
|
ENST00000331872.6
ENST00000311223.5
|
GLUL
|
glutamate-ammonia ligase
|
|
chr5_+_71403061
|
0.237
|
ENST00000512974.1
ENST00000296755.7
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr16_-_87525651
|
0.223
|
ENST00000268616.4
|
ZCCHC14
|
zinc finger, CCHC domain containing 14
|
|
chr2_+_20646824
|
0.212
|
ENST00000272233.4
|
RHOB
|
ras homolog family member B
|
|
chr17_+_55162453
|
0.210
|
ENST00000575322.1
ENST00000337714.3
ENST00000314126.3
|
AKAP1
|
A kinase (PRKA) anchor protein 1
|
|
chrX_+_69509927
|
0.209
|
ENST00000374403.3
|
KIF4A
|
kinesin family member 4A
|
|
chr2_-_166930131
|
0.207
|
ENST00000303395.4
ENST00000409050.1
ENST00000423058.2
ENST00000375405.3
|
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit
|
|
chr12_-_31744031
|
0.200
|
ENST00000389082.5
|
DENND5B
|
DENN/MADD domain containing 5B
|
|
chr15_+_41952591
|
0.200
|
ENST00000566718.1
ENST00000219905.7
ENST00000389936.4
ENST00000545763.1
|
MGA
|
MGA, MAX dimerization protein
|
|
chr16_-_73082274
|
0.199
|
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr13_-_41240717
|
0.187
|
ENST00000379561.5
|
FOXO1
|
forkhead box O1
|
|
chr13_+_50656307
|
0.187
|
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding)
|
|
chr4_-_140098339
|
0.187
|
ENST00000394235.2
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chr10_+_31608054
|
0.185
|
ENST00000320985.10
ENST00000361642.5
ENST00000560721.2
ENST00000558440.1
ENST00000424869.1
ENST00000542815.3
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr17_-_8534067
|
0.184
|
ENST00000360416.3
ENST00000269243.4
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr20_-_4982132
|
0.153
|
ENST00000338244.1
ENST00000424750.2
|
SLC23A2
|
solute carrier family 23 (ascorbic acid transporter), member 2
|
|
chr17_+_16318850
|
0.149
|
ENST00000338560.7
|
TRPV2
|
transient receptor potential cation channel, subfamily V, member 2
|
|
chr11_+_14665263
|
0.145
|
ENST00000282096.4
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited
|
|
chr2_-_174830430
|
0.142
|
ENST00000310015.6
ENST00000455789.2
|
SP3
|
Sp3 transcription factor
|
|
chr6_-_82462425
|
0.141
|
ENST00000369754.3
ENST00000320172.6
ENST00000369756.3
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr18_+_72201829
|
0.135
|
ENST00000582365.1
|
CNDP1
|
carnosine dipeptidase 1 (metallopeptidase M20 family)
|
|
chr4_-_74124502
|
0.134
|
ENST00000358602.4
ENST00000330838.6
ENST00000561029.1
|
ANKRD17
|
ankyrin repeat domain 17
|
|
chr2_+_138721850
|
0.133
|
ENST00000329366.4
ENST00000280097.3
|
HNMT
|
histamine N-methyltransferase
|
|
chr12_-_125348448
|
0.132
|
ENST00000339570.5
|
SCARB1
|
scavenger receptor class B, member 1
|
|
chr19_+_34745442
|
0.129
|
ENST00000299505.6
ENST00000588470.1
ENST00000589583.1
ENST00000588338.2
|
KIAA0355
|
KIAA0355
|
|
chr1_+_65730385
|
0.125
|
ENST00000263441.7
ENST00000395325.3
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr20_-_30795511
|
0.120
|
ENST00000246229.4
|
PLAGL2
|
pleiomorphic adenoma gene-like 2
|
|
chr3_-_13461807
|
0.119
|
ENST00000254508.5
|
NUP210
|
nucleoporin 210kDa
|
|
chr22_+_40573921
|
0.115
|
ENST00000454349.2
ENST00000335727.9
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr7_-_152133059
|
0.110
|
ENST00000262189.6
ENST00000355193.2
|
KMT2C
|
lysine (K)-specific methyltransferase 2C
|
|
chr10_-_97321112
|
0.107
|
ENST00000607232.1
ENST00000371227.4
ENST00000371249.2
ENST00000371247.2
ENST00000371246.2
ENST00000393949.1
ENST00000353505.5
ENST00000347291.4
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr8_-_134584152
|
0.106
|
ENST00000521180.1
ENST00000517668.1
ENST00000319914.5
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1
|
|
chr1_-_190446759
|
0.106
|
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3
|
|
chr5_-_141338627
|
0.101
|
ENST00000231484.3
|
PCDH12
|
protocadherin 12
|
|
chr5_-_132113036
|
0.096
|
ENST00000378706.1
|
SEPT8
|
septin 8
|
|
chr17_-_30669138
|
0.096
|
ENST00000225805.4
ENST00000577809.1
|
C17orf75
|
chromosome 17 open reading frame 75
|
|
chr2_+_159313452
|
0.095
|
ENST00000389757.3
ENST00000389759.3
|
PKP4
|
plakophilin 4
|
|
chr1_+_27022485
|
0.095
|
ENST00000324856.7
|
ARID1A
|
AT rich interactive domain 1A (SWI-like)
|
|
chr17_+_57784826
|
0.092
|
ENST00000262291.4
|
VMP1
|
vacuole membrane protein 1
|
|
chr18_+_60382672
|
0.091
|
ENST00000400316.4
ENST00000262719.5
|
PHLPP1
|
PH domain and leucine rich repeat protein phosphatase 1
|
|
chr1_-_161279749
|
0.090
|
ENST00000533357.1
ENST00000360451.6
ENST00000336559.4
|
MPZ
|
myelin protein zero
|
|
chr1_+_22778337
|
0.088
|
ENST00000404138.1
ENST00000400239.2
ENST00000375647.4
ENST00000374651.4
|
ZBTB40
|
zinc finger and BTB domain containing 40
|
|
chr5_+_65440032
|
0.086
|
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1
|
|
chr2_-_175351744
|
0.085
|
ENST00000295500.4
ENST00000392552.2
ENST00000392551.2
|
GPR155
|
G protein-coupled receptor 155
|
|
chr6_-_34360413
|
0.084
|
ENST00000607016.1
|
NUDT3
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3
|
|
chr3_-_47619623
|
0.084
|
ENST00000456150.1
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C)
|
|
chr1_+_36396677
|
0.083
|
ENST00000373191.4
ENST00000397828.2
|
AGO3
|
argonaute RISC catalytic component 3
|
|
chr11_-_57103327
|
0.080
|
ENST00000529002.1
ENST00000278412.2
|
SSRP1
|
structure specific recognition protein 1
|
|
chr10_-_11653753
|
0.080
|
ENST00000609104.1
|
USP6NL
|
USP6 N-terminal like
|
|
chr6_-_86352642
|
0.077
|
ENST00000355238.6
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr6_+_108881012
|
0.076
|
ENST00000343882.6
|
FOXO3
|
forkhead box O3
|
|
chr5_-_59189545
|
0.076
|
ENST00000340635.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr11_-_73309228
|
0.076
|
ENST00000356467.4
ENST00000064778.4
|
FAM168A
|
family with sequence similarity 168, member A
|
|
chr6_+_155054459
|
0.075
|
ENST00000367178.3
ENST00000417268.1
ENST00000367186.4
|
SCAF8
|
SR-related CTD-associated factor 8
|
|
chr20_-_32274179
|
0.072
|
ENST00000343380.5
|
E2F1
|
E2F transcription factor 1
|
|
chr2_+_99953816
|
0.072
|
ENST00000289371.6
|
EIF5B
|
eukaryotic translation initiation factor 5B
|
|
chr15_-_49447835
|
0.071
|
ENST00000388901.5
ENST00000299259.6
|
COPS2
|
COP9 signalosome subunit 2
|
|
chr2_+_24714729
|
0.071
|
ENST00000406961.1
ENST00000405141.1
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr2_+_29117509
|
0.069
|
ENST00000407426.3
|
WDR43
|
WD repeat domain 43
|
|
chr2_+_135676381
|
0.069
|
ENST00000537343.1
ENST00000295238.6
ENST00000264157.5
|
CCNT2
|
cyclin T2
|
|
chr1_+_97187318
|
0.068
|
ENST00000609116.1
ENST00000370198.1
ENST00000370197.1
ENST00000426398.2
ENST00000394184.3
|
PTBP2
|
polypyrimidine tract binding protein 2
|
|
chr3_-_72496035
|
0.065
|
ENST00000477973.2
|
RYBP
|
RING1 and YY1 binding protein
|
|
chr5_+_145826867
|
0.063
|
ENST00000296702.5
ENST00000394421.2
|
TCERG1
|
transcription elongation regulator 1
|
|
chr6_+_106546808
|
0.062
|
ENST00000369089.3
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr2_-_69614373
|
0.062
|
ENST00000361060.5
ENST00000357308.4
|
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1
|
|
chr2_-_40679186
|
0.062
|
ENST00000406785.2
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr1_+_167190066
|
0.061
|
ENST00000367866.2
ENST00000429375.2
ENST00000452019.1
ENST00000420254.3
ENST00000541643.3
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr7_-_105517021
|
0.061
|
ENST00000318724.4
ENST00000419735.3
|
ATXN7L1
|
ataxin 7-like 1
|
|
chrX_-_100307076
|
0.060
|
ENST00000338687.7
ENST00000545398.1
ENST00000372931.5
|
TRMT2B
|
tRNA methyltransferase 2 homolog B (S. cerevisiae)
|
|
chr1_-_205719295
|
0.060
|
ENST00000367142.4
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1
|
|
chr12_+_111843749
|
0.060
|
ENST00000341259.2
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr6_+_69345166
|
0.055
|
ENST00000370598.1
|
BAI3
|
brain-specific angiogenesis inhibitor 3
|
|
chr16_+_2587998
|
0.055
|
ENST00000441549.3
ENST00000268673.7
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr16_+_19422035
|
0.054
|
ENST00000381414.4
ENST00000396229.2
|
TMC5
|
transmembrane channel-like 5
|
|
chr10_-_101989315
|
0.053
|
ENST00000370397.7
|
CHUK
|
conserved helix-loop-helix ubiquitous kinase
|
|
chr15_+_77223960
|
0.051
|
ENST00000394885.3
|
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain
|
|
chr2_-_240322643
|
0.051
|
ENST00000345617.3
|
HDAC4
|
histone deacetylase 4
|
|
chr1_+_118148556
|
0.050
|
ENST00000369448.3
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr2_-_211036051
|
0.050
|
ENST00000418791.1
ENST00000452086.1
ENST00000281772.9
|
KANSL1L
|
KAT8 regulatory NSL complex subunit 1-like
|
|
chr10_-_94003003
|
0.050
|
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr3_+_142720366
|
0.049
|
ENST00000493782.1
ENST00000397933.2
ENST00000473835.2
ENST00000493598.2
|
U2SURP
|
U2 snRNP-associated SURP domain containing
|
|
chr16_+_48278178
|
0.049
|
ENST00000285737.4
ENST00000535754.1
|
LONP2
|
lon peptidase 2, peroxisomal
|
|
chr1_-_205601064
|
0.046
|
ENST00000357992.4
ENST00000289703.4
|
ELK4
|
ELK4, ETS-domain protein (SRF accessory protein 1)
|
|
chr1_-_153935983
|
0.046
|
ENST00000537590.1
ENST00000356205.4
|
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1
|
|
chr11_+_119019722
|
0.042
|
ENST00000307417.3
|
ABCG4
|
ATP-binding cassette, sub-family G (WHITE), member 4
|
|
chr16_-_58663720
|
0.041
|
ENST00000564557.1
ENST00000569240.1
ENST00000441024.2
ENST00000569020.1
ENST00000317147.5
|
CNOT1
|
CCR4-NOT transcription complex, subunit 1
|
|
chr11_-_31839488
|
0.041
|
ENST00000419022.1
ENST00000379132.3
ENST00000379129.2
|
PAX6
|
paired box 6
|
|
chr13_+_33160553
|
0.041
|
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae)
|
|
chr8_-_41909496
|
0.040
|
ENST00000265713.2
ENST00000406337.1
ENST00000396930.3
ENST00000485568.1
ENST00000426524.1
|
KAT6A
|
K(lysine) acetyltransferase 6A
|
|
chr12_+_70760056
|
0.039
|
ENST00000258111.4
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chr3_-_126194707
|
0.038
|
ENST00000336332.5
ENST00000389709.3
|
ZXDC
|
ZXD family zinc finger C
|
|
chr1_-_70671216
|
0.037
|
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40
|
|
chr19_+_49375649
|
0.037
|
ENST00000200453.5
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A
|
|
chr7_-_15601595
|
0.036
|
ENST00000342526.3
|
AGMO
|
alkylglycerol monooxygenase
|
|
chrX_+_24167746
|
0.035
|
ENST00000428571.1
ENST00000539115.1
|
ZFX
|
zinc finger protein, X-linked
|
|
chrX_+_77166172
|
0.034
|
ENST00000343533.5
ENST00000350425.4
ENST00000341514.6
|
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide
|
|
chr3_+_152017181
|
0.032
|
ENST00000498502.1
ENST00000324196.5
ENST00000545754.1
ENST00000357472.3
|
MBNL1
|
muscleblind-like splicing regulator 1
|
|
chr1_-_38397384
|
0.032
|
ENST00000373027.1
|
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa
|
|
chr1_+_247579451
|
0.032
|
ENST00000391828.3
ENST00000366497.2
|
NLRP3
|
NLR family, pyrin domain containing 3
|
|
chr9_+_133454943
|
0.031
|
ENST00000319725.9
|
FUBP3
|
far upstream element (FUSE) binding protein 3
|
|
chr1_+_151043070
|
0.030
|
ENST00000368918.3
ENST00000368917.1
|
GABPB2
|
GA binding protein transcription factor, beta subunit 2
|
|
chr13_+_24734844
|
0.030
|
ENST00000382108.3
|
SPATA13
|
spermatogenesis associated 13
|
|
chr15_-_68521996
|
0.028
|
ENST00000418702.2
ENST00000565471.1
ENST00000564752.1
ENST00000566347.1
ENST00000249806.5
ENST00000562767.1
|
CLN6
RP11-315D16.2
|
ceroid-lipofuscinosis, neuronal 6, late infantile, variant
Uncharacterized protein
|
|
chr20_-_33999766
|
0.027
|
ENST00000349714.5
ENST00000438533.1
ENST00000359226.2
ENST00000374384.2
ENST00000374377.5
ENST00000407996.2
ENST00000424405.1
ENST00000542501.1
ENST00000397554.1
ENST00000540457.1
ENST00000374380.2
ENST00000374385.5
|
UQCC1
|
ubiquinol-cytochrome c reductase complex assembly factor 1
|
|
chr2_+_204192942
|
0.026
|
ENST00000295851.5
ENST00000261017.5
|
ABI2
|
abl-interactor 2
|
|
chr12_+_104324112
|
0.026
|
ENST00000299767.5
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1
|
|
chr2_+_166095898
|
0.026
|
ENST00000424833.1
ENST00000375437.2
ENST00000357398.3
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr1_+_180601139
|
0.025
|
ENST00000367590.4
ENST00000367589.3
|
XPR1
|
xenotropic and polytropic retrovirus receptor 1
|
|
chr17_+_57970469
|
0.024
|
ENST00000443572.2
ENST00000406116.3
ENST00000225577.4
ENST00000393021.3
|
RPS6KB1
|
ribosomal protein S6 kinase, 70kDa, polypeptide 1
|
|
chr3_+_172468472
|
0.023
|
ENST00000232458.5
ENST00000392692.3
|
ECT2
|
epithelial cell transforming sequence 2 oncogene
|
|
chr17_-_4167142
|
0.022
|
ENST00000570535.1
ENST00000574367.1
ENST00000341657.4
ENST00000433651.1
|
ANKFY1
|
ankyrin repeat and FYVE domain containing 1
|
|
chr15_-_64648273
|
0.021
|
ENST00000607537.1
ENST00000303052.7
ENST00000303032.6
|
CSNK1G1
|
casein kinase 1, gamma 1
|
|
chr11_-_62572901
|
0.021
|
ENST00000439713.2
ENST00000531131.1
ENST00000530875.1
ENST00000531709.2
ENST00000294172.2
|
NXF1
|
nuclear RNA export factor 1
|
|
chr21_+_44394620
|
0.020
|
ENST00000291547.5
|
PKNOX1
|
PBX/knotted 1 homeobox 1
|
|
chr1_-_39339777
|
0.020
|
ENST00000397572.2
|
MYCBP
|
MYC binding protein
|
|
chr11_+_73661364
|
0.019
|
ENST00000339764.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13
|
|
chr6_+_43484760
|
0.019
|
ENST00000372389.3
ENST00000372344.2
ENST00000304004.3
ENST00000423780.1
|
POLR1C
|
polymerase (RNA) I polypeptide C, 30kDa
|
|
chr1_-_108507631
|
0.019
|
ENST00000527011.1
ENST00000370056.4
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr11_-_95657231
|
0.018
|
ENST00000409459.1
ENST00000352297.7
ENST00000393223.3
ENST00000346299.5
|
MTMR2
|
myotubularin related protein 2
|
|
chr6_-_99395787
|
0.018
|
ENST00000369244.2
ENST00000229971.1
|
FBXL4
|
F-box and leucine-rich repeat protein 4
|
|
chr21_+_30396950
|
0.017
|
ENST00000399975.3
ENST00000399976.2
ENST00000334352.4
ENST00000399973.1
ENST00000535828.1
|
USP16
|
ubiquitin specific peptidase 16
|
|
chr16_-_20911641
|
0.017
|
ENST00000324344.4
ENST00000564349.1
|
DCUN1D3
ERI2
|
DCN1, defective in cullin neddylation 1, domain containing 3
ERI1 exoribonuclease family member 2
|
|
chr2_+_45878790
|
0.016
|
ENST00000306156.3
|
PRKCE
|
protein kinase C, epsilon
|
|
chr7_+_107110488
|
0.014
|
ENST00000304402.4
|
GPR22
|
G protein-coupled receptor 22
|
|
chr8_-_66546439
|
0.014
|
ENST00000276569.3
|
ARMC1
|
armadillo repeat containing 1
|
|
chr17_-_8066250
|
0.014
|
ENST00000488857.1
ENST00000481878.1
ENST00000316509.6
ENST00000498285.1
|
VAMP2
RP11-599B13.6
|
vesicle-associated membrane protein 2 (synaptobrevin 2)
Uncharacterized protein
|
|
chr7_-_44924939
|
0.014
|
ENST00000395699.2
|
PURB
|
purine-rich element binding protein B
|
|
chr12_+_70636765
|
0.013
|
ENST00000552231.1
ENST00000229195.3
ENST00000547780.1
ENST00000418359.3
|
CNOT2
|
CCR4-NOT transcription complex, subunit 2
|
|
chr1_-_109968973
|
0.013
|
ENST00000271308.4
ENST00000538610.1
|
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5
|
|
chr7_-_123389104
|
0.009
|
ENST00000223023.4
|
WASL
|
Wiskott-Aldrich syndrome-like
|
|
chr2_+_99061298
|
0.009
|
ENST00000409016.4
ENST00000409851.3
ENST00000409463.1
ENST00000074304.5
ENST00000545415.1
ENST00000409540.3
|
INPP4A
|
inositol polyphosphate-4-phosphatase, type I, 107kDa
|
|
chr16_+_50187556
|
0.008
|
ENST00000561678.1
ENST00000357464.3
|
PAPD5
|
PAP associated domain containing 5
|
|
chr13_-_41706864
|
0.007
|
ENST00000379485.1
ENST00000499385.2
|
KBTBD6
|
kelch repeat and BTB (POZ) domain containing 6
|
|
chr11_-_132813566
|
0.006
|
ENST00000331898.7
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr1_+_244214577
|
0.006
|
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18
|
|
chr12_-_90049828
|
0.006
|
ENST00000261173.2
ENST00000348959.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr3_-_105587879
|
0.005
|
ENST00000264122.4
ENST00000403724.1
ENST00000405772.1
|
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase
|
|
chrX_+_16804544
|
0.005
|
ENST00000380122.5
ENST00000398155.4
|
TXLNG
|
taxilin gamma
|
|
chr15_-_52861394
|
0.004
|
ENST00000563277.1
ENST00000566423.1
|
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa
|
|
chr15_-_49338748
|
0.004
|
ENST00000559471.1
|
SECISBP2L
|
SECIS binding protein 2-like
|
|
chr6_-_28367510
|
0.004
|
ENST00000361028.1
|
ZSCAN12
|
zinc finger and SCAN domain containing 12
|
|
chr8_-_89339705
|
0.003
|
ENST00000286614.6
|
MMP16
|
matrix metallopeptidase 16 (membrane-inserted)
|
|
chrX_+_16737718
|
0.003
|
ENST00000380155.3
|
SYAP1
|
synapse associated protein 1
|
|
chr8_-_74791051
|
0.003
|
ENST00000453587.2
ENST00000602969.1
ENST00000602593.1
ENST00000419880.3
ENST00000517608.1
|
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative)
|
|
chr1_-_28415204
|
0.002
|
ENST00000373871.3
|
EYA3
|
eyes absent homolog 3 (Drosophila)
|
|
chr16_+_67880574
|
0.001
|
ENST00000219169.4
|
NUTF2
|
nuclear transport factor 2
|