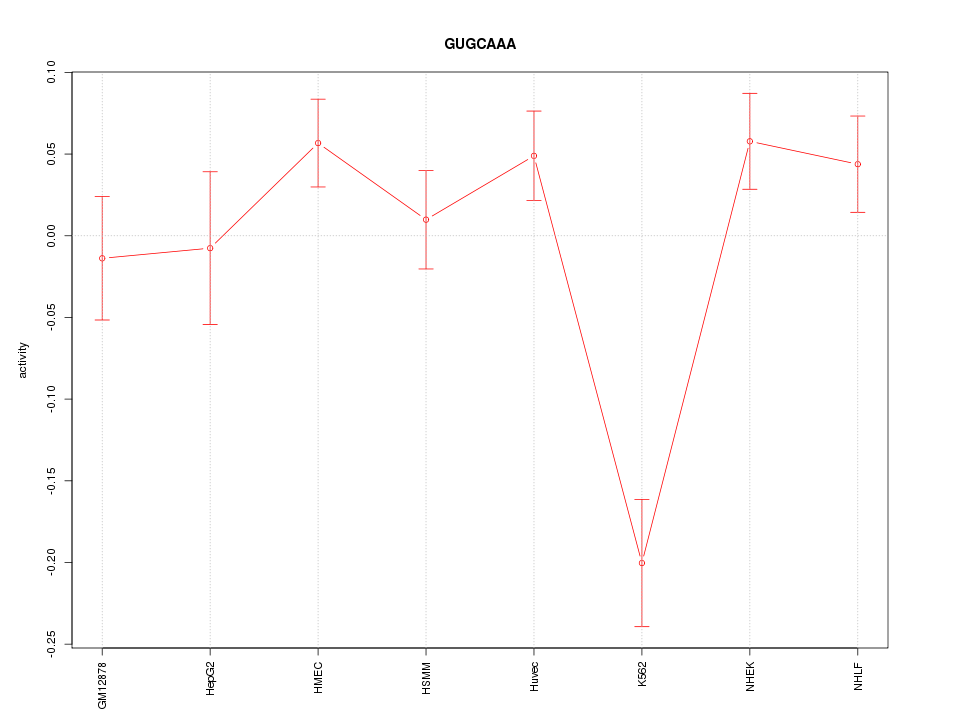
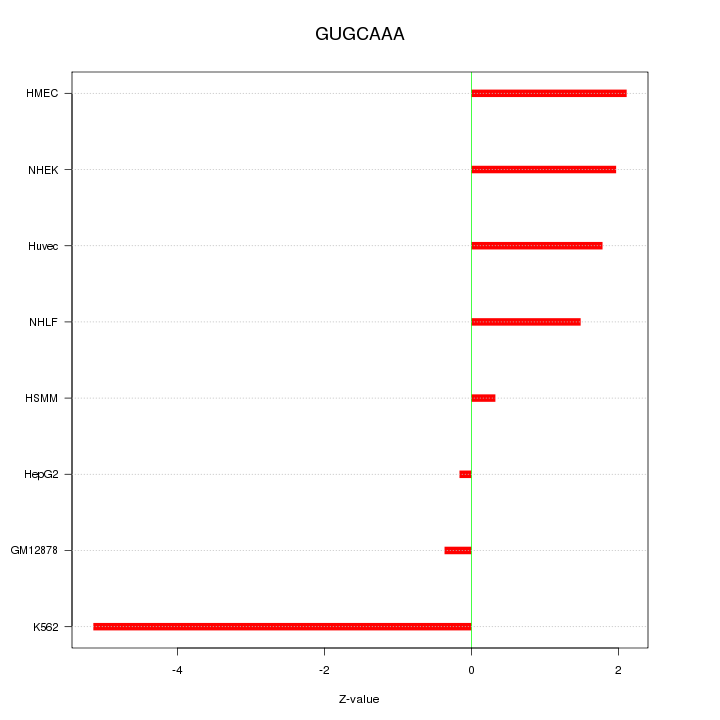
Motif ID: GUGCAAA
Z-value: 2.249

Mature miRNA associated with seed GUGCAAA:
| Name | miRBase Accession |
|---|---|
| hsa-miR-19a-3p | MIMAT0000073 |
| hsa-miR-19b-3p | MIMAT0000074 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 1.1 | 4.6 | GO:0051503 | purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) |
| 1.1 | 3.2 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 1.1 | 3.2 | GO:2000064 | regulation of cortisol biosynthetic process(GO:2000064) |
| 1.0 | 2.9 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) intestinal epithelial cell development(GO:0060576) |
| 0.9 | 4.3 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.8 | 2.4 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.7 | 2.7 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.6 | 3.2 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.6 | 1.7 | GO:0048627 | myoblast development(GO:0048627) |
| 0.5 | 2.6 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.5 | 1.6 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.5 | 2.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.5 | 1.5 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.5 | 1.4 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.4 | 1.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.4 | 1.1 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.4 | 1.5 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.3 | 2.0 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.3 | 2.0 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.3 | 0.8 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.3 | 0.8 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.2 | 1.7 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 | 3.8 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.2 | 0.7 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 0.7 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.2 | 0.7 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.2 | 1.3 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.2 | 2.0 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.2 | 0.6 | GO:0034139 | regulation of toll-like receptor 2 signaling pathway(GO:0034135) regulation of toll-like receptor 3 signaling pathway(GO:0034139) negative regulation of epithelial cell apoptotic process(GO:1904036) negative regulation of endothelial cell apoptotic process(GO:2000352) |
| 0.2 | 1.8 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.2 | 5.0 | GO:0001960 | negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.2 | 0.6 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.2 | 0.9 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.2 | 0.5 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 0.6 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.1 | 0.7 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 0.5 | GO:1903530 | regulation of secretion by cell(GO:1903530) |
| 0.1 | 1.6 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 1.2 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 0.8 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.1 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.1 | 0.2 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 0.3 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.1 | 0.6 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.1 | 0.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.4 | GO:0034204 | Cdc42 protein signal transduction(GO:0032488) lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 0.3 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.1 | 0.3 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 2.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.1 | 1.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.9 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 0.7 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 0.5 | GO:0032472 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) Golgi calcium ion transport(GO:0032472) manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.9 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.2 | GO:0046449 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.1 | 0.2 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 0.8 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 0.3 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.7 | GO:0007409 | axonogenesis(GO:0007409) |
| 0.1 | 1.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.6 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 0.2 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.1 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 0.4 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 0.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.9 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 1.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.3 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.1 | 0.6 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 | 0.4 | GO:0000042 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 1.0 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.1 | 0.8 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 0.4 | GO:0071731 | interleukin-1-mediated signaling pathway(GO:0070498) response to nitric oxide(GO:0071731) |
| 0.0 | 2.0 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.9 | GO:0007507 | heart development(GO:0007507) |
| 0.0 | 0.5 | GO:0046051 | GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.0 | 0.3 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.5 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.8 | GO:0021772 | olfactory bulb development(GO:0021772) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.2 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.6 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.6 | GO:0060438 | trachea development(GO:0060438) |
| 0.0 | 0.1 | GO:0046732 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by virus of host immune response(GO:0075528) |
| 0.0 | 0.2 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 2.8 | GO:0034332 | adherens junction organization(GO:0034332) |
| 0.0 | 0.3 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 | 0.3 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.7 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 1.1 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.1 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) regulation of protein folding(GO:1903332) negative regulation of protein folding(GO:1903333) |
| 0.0 | 0.4 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.2 | GO:0045060 | negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.3 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.2 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.3 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.5 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.3 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.8 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.1 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.5 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 1.4 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 1.6 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) negative regulation of cell projection organization(GO:0031345) |
| 0.0 | 4.2 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 0.1 | GO:0006896 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.4 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 0.9 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.2 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 4.3 | GO:0030522 | intracellular receptor signaling pathway(GO:0030522) |
| 0.0 | 0.6 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.0 | 0.5 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.2 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.5 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 1.4 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0072148 | epithelial cell fate commitment(GO:0072148) |
| 0.0 | 0.0 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.7 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.0 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 | 0.2 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.6 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.2 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0060678 | dichotomous subdivision of terminal units involved in ureteric bud branching(GO:0060678) |
| 0.0 | 0.5 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.0 | 0.1 | GO:0048875 | positive regulation of synaptic transmission, GABAergic(GO:0032230) surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.4 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 3.0 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.2 | GO:0090174 | organelle membrane fusion(GO:0090174) |
| 0.0 | 0.0 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.0 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.4 | 4.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 4.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.4 | 2.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.4 | 2.4 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.3 | 1.6 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.2 | 4.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 1.7 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.6 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 1.5 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 0.4 | GO:0000806 | Y chromosome(GO:0000806) |
| 0.1 | 0.9 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 3.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.6 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 4.8 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 0.4 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 1.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 1.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 1.1 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.4 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.0 | GO:0090568 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.4 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 2.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 2.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 2.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.0 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 7.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.7 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 1.5 | 4.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.6 | 1.9 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.6 | 3.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.6 | 2.4 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.6 | 2.3 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) protein phosphatase activator activity(GO:0072542) |
| 0.5 | 1.6 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.5 | 1.9 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.5 | 1.4 | GO:0048184 | obsolete follistatin binding(GO:0048184) |
| 0.4 | 4.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.4 | 4.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.4 | 4.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.4 | 1.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.4 | 1.4 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.3 | 1.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 0.8 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.2 | 0.7 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.2 | 0.7 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.2 | 0.9 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.2 | 1.5 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 2.0 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.2 | 3.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 2.2 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.2 | 1.0 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 1.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 1.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.5 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 0.7 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.1 | 0.4 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.4 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.3 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 0.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.5 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 1.5 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.1 | 0.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 1.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 2.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 1.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 1.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 1.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.8 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.8 | GO:0046625 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) sphingolipid binding(GO:0046625) |
| 0.1 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.5 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 0.8 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.1 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 1.3 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 1.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 2.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.7 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.7 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.4 | GO:0016944 | obsolete RNA polymerase II transcription elongation factor activity(GO:0016944) |
| 0.0 | 0.9 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.5 | GO:0004691 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.1 | GO:0015217 | purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) ATP:ADP antiporter activity(GO:0005471) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.6 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.2 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.9 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 1.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 4.0 | GO:0016564 | obsolete transcription repressor activity(GO:0016564) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 1.1 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.2 | GO:0004673 | phosphorelay sensor kinase activity(GO:0000155) protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.0 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.2 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 2.5 | GO:0008022 | protein C-terminus binding(GO:0008022) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.4 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.4 | ST_PAC1_RECEPTOR_PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.2 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.2 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |