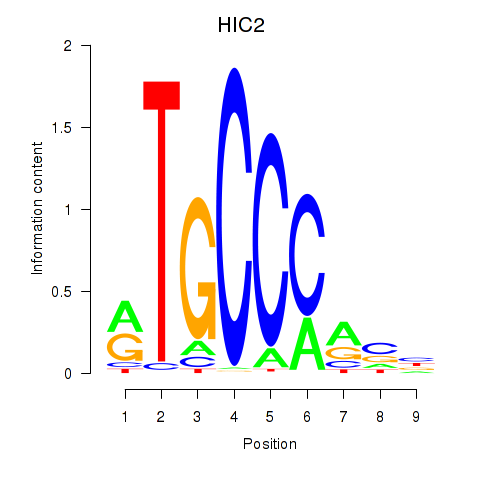
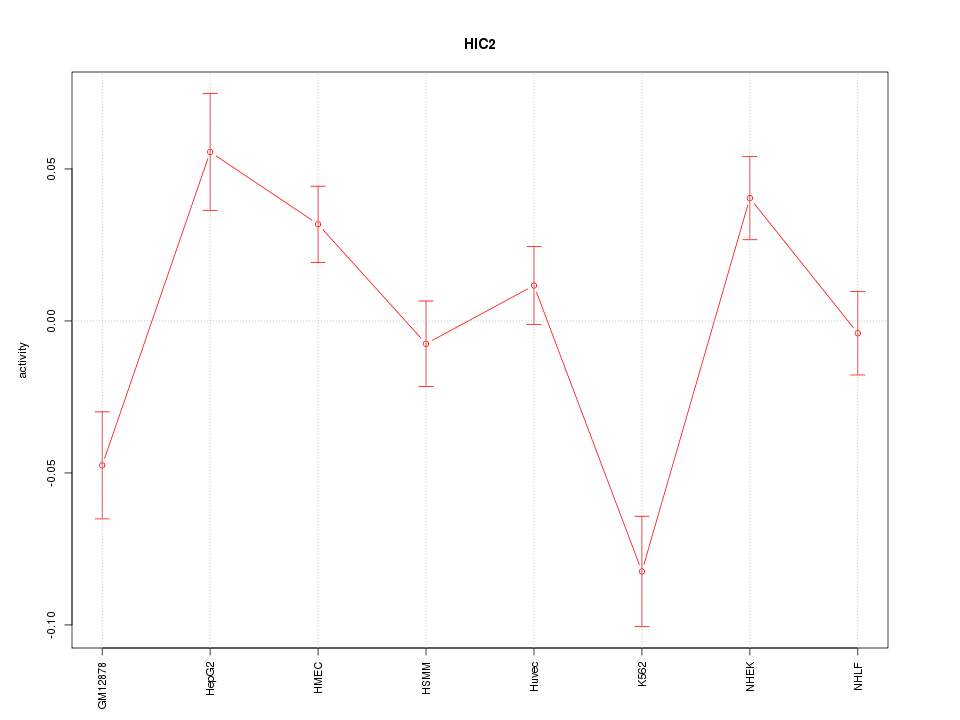
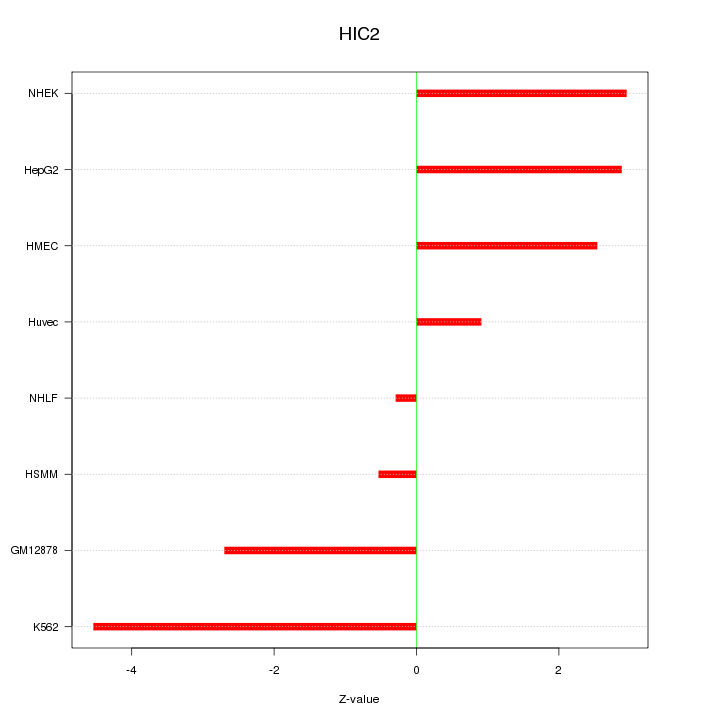
| 3.4 |
13.5 |
GO:0033986 |
response to methanol(GO:0033986) response to chromate(GO:0046687) |
| 2.0 |
6.0 |
GO:0010273 |
detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 1.5 |
4.5 |
GO:0071603 |
negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) endothelial cell-cell adhesion(GO:0071603) |
| 1.3 |
3.9 |
GO:0010982 |
regulation of high-density lipoprotein particle clearance(GO:0010982) regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 1.1 |
3.4 |
GO:0010757 |
negative regulation of plasminogen activation(GO:0010757) |
| 0.9 |
24.9 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.8 |
2.5 |
GO:0006015 |
5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.8 |
3.0 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.7 |
3.6 |
GO:0044854 |
membrane raft assembly(GO:0001765) nitric oxide homeostasis(GO:0033484) negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) membrane assembly(GO:0071709) |
| 0.7 |
3.5 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.6 |
1.9 |
GO:0010902 |
positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.6 |
6.0 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.6 |
3.0 |
GO:0045989 |
positive regulation of striated muscle contraction(GO:0045989) |
| 0.6 |
1.8 |
GO:0048627 |
myoblast development(GO:0048627) |
| 0.6 |
5.6 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.6 |
1.7 |
GO:0021836 |
chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.5 |
2.0 |
GO:0030913 |
paranodal junction assembly(GO:0030913) |
| 0.5 |
1.5 |
GO:0061117 |
negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.5 |
1.4 |
GO:0051541 |
elastin metabolic process(GO:0051541) |
| 0.4 |
1.3 |
GO:0060729 |
negative regulation of interleukin-23 production(GO:0032707) intestinal epithelial structure maintenance(GO:0060729) |
| 0.4 |
1.3 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.4 |
1.3 |
GO:0032488 |
Cdc42 protein signal transduction(GO:0032488) |
| 0.4 |
1.2 |
GO:0033240 |
positive regulation of cellular amine metabolic process(GO:0033240) |
| 0.4 |
1.2 |
GO:0002689 |
negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.4 |
2.2 |
GO:0032460 |
negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.4 |
1.1 |
GO:0021754 |
facial nucleus development(GO:0021754) |
| 0.4 |
1.4 |
GO:0042268 |
regulation of cytolysis(GO:0042268) |
| 0.4 |
2.1 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.3 |
1.0 |
GO:0060574 |
trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) intestinal epithelial cell maturation(GO:0060574) intestinal epithelial cell development(GO:0060576) |
| 0.3 |
0.9 |
GO:0002575 |
basophil chemotaxis(GO:0002575) |
| 0.3 |
1.2 |
GO:0006701 |
progesterone biosynthetic process(GO:0006701) |
| 0.3 |
1.8 |
GO:0031639 |
plasminogen activation(GO:0031639) |
| 0.3 |
1.8 |
GO:0042420 |
dopamine catabolic process(GO:0042420) |
| 0.3 |
0.9 |
GO:0048389 |
intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) bud dilation involved in lung branching(GO:0060503) branch elongation involved in ureteric bud branching(GO:0060681) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) pulmonary artery morphogenesis(GO:0061156) cell proliferation involved in mesonephros development(GO:0061209) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) negative regulation of mesonephros development(GO:0061218) pattern specification involved in mesonephros development(GO:0061227) renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) BMP signaling pathway involved in nephric duct formation(GO:0071893) glomerular visceral epithelial cell development(GO:0072015) comma-shaped body morphogenesis(GO:0072049) regulation of branch elongation involved in ureteric bud branching(GO:0072095) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) glomerulus morphogenesis(GO:0072102) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) nephric duct formation(GO:0072179) ureter urothelium development(GO:0072190) ureter epithelial cell differentiation(GO:0072192) ureter morphogenesis(GO:0072197) metanephric comma-shaped body morphogenesis(GO:0072278) glomerular epithelial cell development(GO:0072310) negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.3 |
2.3 |
GO:1904893 |
negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.3 |
0.3 |
GO:1904036 |
negative regulation of epithelial cell apoptotic process(GO:1904036) negative regulation of endothelial cell apoptotic process(GO:2000352) |
| 0.3 |
1.8 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.3 |
1.0 |
GO:0045715 |
negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.3 |
0.5 |
GO:0050918 |
positive chemotaxis(GO:0050918) |
| 0.3 |
1.3 |
GO:0019471 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.3 |
0.8 |
GO:0045719 |
negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.2 |
0.7 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 |
1.4 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.2 |
0.9 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.2 |
0.9 |
GO:0060649 |
nipple development(GO:0060618) mammary gland bud elongation(GO:0060649) nipple morphogenesis(GO:0060658) nipple sheath formation(GO:0060659) |
| 0.2 |
0.7 |
GO:0002124 |
territorial aggressive behavior(GO:0002124) brainstem development(GO:0003360) vocalization behavior(GO:0071625) |
| 0.2 |
3.4 |
GO:0048873 |
homeostasis of number of cells within a tissue(GO:0048873) |
| 0.2 |
1.7 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 |
1.3 |
GO:0051387 |
negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.2 |
0.4 |
GO:0021783 |
preganglionic parasympathetic fiber development(GO:0021783) |
| 0.2 |
0.6 |
GO:0008206 |
bile acid metabolic process(GO:0008206) |
| 0.2 |
0.6 |
GO:1903961 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.2 |
0.6 |
GO:0071109 |
superior temporal gyrus development(GO:0071109) |
| 0.2 |
1.2 |
GO:0048010 |
vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.2 |
3.0 |
GO:0050872 |
white fat cell differentiation(GO:0050872) |
| 0.2 |
1.4 |
GO:0032000 |
positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 |
0.6 |
GO:0050917 |
sensory perception of umami taste(GO:0050917) |
| 0.2 |
0.2 |
GO:0001702 |
gastrulation with mouth forming second(GO:0001702) |
| 0.2 |
3.0 |
GO:0032456 |
endocytic recycling(GO:0032456) |
| 0.2 |
0.2 |
GO:0060677 |
ureteric bud elongation(GO:0060677) |
| 0.2 |
0.4 |
GO:0051088 |
PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.2 |
1.0 |
GO:0009439 |
cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.2 |
2.6 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.2 |
1.5 |
GO:0045542 |
positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.2 |
2.0 |
GO:0002544 |
chronic inflammatory response(GO:0002544) |
| 0.2 |
0.4 |
GO:0032914 |
positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.2 |
1.1 |
GO:0030041 |
actin filament polymerization(GO:0030041) |
| 0.2 |
3.9 |
GO:0030514 |
negative regulation of BMP signaling pathway(GO:0030514) |
| 0.2 |
0.9 |
GO:0006290 |
pyrimidine dimer repair(GO:0006290) |
| 0.2 |
1.2 |
GO:0060157 |
urinary bladder development(GO:0060157) |
| 0.2 |
0.5 |
GO:0007614 |
short-term memory(GO:0007614) |
| 0.2 |
1.2 |
GO:0097384 |
cellular lipid biosynthetic process(GO:0097384) |
| 0.2 |
0.7 |
GO:2000114 |
regulation of establishment of cell polarity(GO:2000114) |
| 0.2 |
0.8 |
GO:0060414 |
aorta development(GO:0035904) aorta morphogenesis(GO:0035909) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 |
0.5 |
GO:0034447 |
very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.2 |
1.8 |
GO:0045741 |
positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.2 |
0.9 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.2 |
1.4 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 |
0.4 |
GO:0052805 |
histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.1 |
0.9 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 |
0.4 |
GO:0070672 |
response to interleukin-15(GO:0070672) |
| 0.1 |
2.8 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.1 |
0.9 |
GO:0000098 |
sulfur amino acid catabolic process(GO:0000098) |
| 0.1 |
0.1 |
GO:0032672 |
regulation of interleukin-3 production(GO:0032672) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) |
| 0.1 |
0.3 |
GO:0045103 |
intermediate filament-based process(GO:0045103) |
| 0.1 |
0.3 |
GO:0021592 |
fourth ventricle development(GO:0021592) |
| 0.1 |
1.2 |
GO:0051552 |
flavone metabolic process(GO:0051552) |
| 0.1 |
0.4 |
GO:0019367 |
fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.1 |
2.2 |
GO:0001960 |
negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.1 |
0.1 |
GO:0009414 |
response to water deprivation(GO:0009414) response to water(GO:0009415) |
| 0.1 |
0.8 |
GO:0032007 |
negative regulation of TOR signaling(GO:0032007) |
| 0.1 |
0.8 |
GO:0050919 |
negative chemotaxis(GO:0050919) |
| 0.1 |
0.4 |
GO:0048319 |
axial mesoderm morphogenesis(GO:0048319) |
| 0.1 |
0.4 |
GO:0003289 |
cardiac right ventricle morphogenesis(GO:0003215) septum primum development(GO:0003284) atrial septum primum morphogenesis(GO:0003289) |
| 0.1 |
0.4 |
GO:0061687 |
detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.1 |
1.4 |
GO:0034435 |
steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.1 |
0.9 |
GO:0016199 |
axon midline choice point recognition(GO:0016199) |
| 0.1 |
1.8 |
GO:0034446 |
substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 |
0.5 |
GO:0045349 |
interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 |
1.0 |
GO:0055003 |
cardiac myofibril assembly(GO:0055003) |
| 0.1 |
1.1 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 |
1.2 |
GO:0046653 |
tetrahydrofolate metabolic process(GO:0046653) |
| 0.1 |
0.2 |
GO:0051928 |
positive regulation of calcium ion transport(GO:0051928) |
| 0.1 |
0.5 |
GO:2001258 |
negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) negative regulation of calcium ion transmembrane transporter activity(GO:1901020) negative regulation of cation channel activity(GO:2001258) |
| 0.1 |
0.9 |
GO:0006553 |
lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.1 |
0.7 |
GO:0043045 |
DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.1 |
0.2 |
GO:0060014 |
granulosa cell differentiation(GO:0060014) |
| 0.1 |
0.1 |
GO:0003307 |
regulation of Wnt signaling pathway involved in heart development(GO:0003307) |
| 0.1 |
0.3 |
GO:0031023 |
microtubule organizing center organization(GO:0031023) |
| 0.1 |
0.2 |
GO:0060896 |
neural plate anterior/posterior regionalization(GO:0021999) neural plate pattern specification(GO:0060896) neural plate regionalization(GO:0060897) |
| 0.1 |
0.3 |
GO:0001757 |
somite specification(GO:0001757) |
| 0.1 |
0.3 |
GO:0070254 |
mucus secretion(GO:0070254) |
| 0.1 |
0.5 |
GO:0090343 |
positive regulation of cell aging(GO:0090343) |
| 0.1 |
0.2 |
GO:0046060 |
dATP metabolic process(GO:0046060) |
| 0.1 |
0.2 |
GO:0051597 |
response to methylmercury(GO:0051597) |
| 0.1 |
0.7 |
GO:0032331 |
negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 |
0.3 |
GO:0042369 |
vitamin D catabolic process(GO:0042369) |
| 0.1 |
0.4 |
GO:0042866 |
pyruvate biosynthetic process(GO:0042866) |
| 0.1 |
2.4 |
GO:0007398 |
ectoderm development(GO:0007398) |
| 0.1 |
0.3 |
GO:0006655 |
phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.1 |
0.8 |
GO:0044341 |
sodium-dependent phosphate transport(GO:0044341) |
| 0.1 |
0.6 |
GO:0051665 |
membrane raft localization(GO:0051665) |
| 0.1 |
0.5 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 |
0.8 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.1 |
1.0 |
GO:0019885 |
antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 |
0.6 |
GO:0016559 |
peroxisome fission(GO:0016559) |
| 0.1 |
0.3 |
GO:0071374 |
cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.1 |
1.1 |
GO:0051608 |
histamine transport(GO:0051608) |
| 0.1 |
0.6 |
GO:0006896 |
Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.1 |
0.6 |
GO:0010224 |
response to UV-B(GO:0010224) |
| 0.1 |
1.1 |
GO:0046697 |
decidualization(GO:0046697) |
| 0.1 |
0.3 |
GO:0060839 |
radial pattern formation(GO:0009956) lymphatic endothelial cell fate commitment(GO:0060838) endothelial cell fate commitment(GO:0060839) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 |
0.4 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.1 |
0.4 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) |
| 0.1 |
1.2 |
GO:0060056 |
mammary gland involution(GO:0060056) |
| 0.1 |
0.2 |
GO:0035020 |
regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 |
0.3 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.1 |
0.2 |
GO:0034036 |
purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 |
0.5 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.1 |
0.1 |
GO:0090342 |
regulation of cell aging(GO:0090342) |
| 0.1 |
0.4 |
GO:2000124 |
cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 |
1.2 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.1 |
0.2 |
GO:0045079 |
negative regulation of chemokine production(GO:0032682) negative regulation of chemokine biosynthetic process(GO:0045079) neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) |
| 0.1 |
0.7 |
GO:0010633 |
negative regulation of epithelial cell migration(GO:0010633) |
| 0.1 |
0.4 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 |
0.3 |
GO:0032341 |
aldosterone metabolic process(GO:0032341) aldosterone biosynthetic process(GO:0032342) |
| 0.1 |
0.3 |
GO:0018106 |
peptidyl-histidine phosphorylation(GO:0018106) |
| 0.1 |
0.4 |
GO:0070120 |
ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.1 |
0.2 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 |
0.2 |
GO:0060454 |
positive regulation of gastric acid secretion(GO:0060454) regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.1 |
0.8 |
GO:0007130 |
synaptonemal complex assembly(GO:0007130) |
| 0.1 |
0.5 |
GO:0006700 |
C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 |
0.5 |
GO:0015904 |
tetracycline transport(GO:0015904) |
| 0.1 |
0.5 |
GO:0010658 |
striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 |
0.7 |
GO:0050435 |
beta-amyloid metabolic process(GO:0050435) |
| 0.1 |
0.7 |
GO:0060216 |
definitive hemopoiesis(GO:0060216) |
| 0.1 |
0.3 |
GO:0042905 |
9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 |
0.1 |
GO:0007182 |
common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.1 |
0.2 |
GO:0014831 |
intestine smooth muscle contraction(GO:0014827) gastro-intestinal system smooth muscle contraction(GO:0014831) |
| 0.1 |
0.4 |
GO:0048296 |
isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 |
0.6 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 |
0.2 |
GO:0043162 |
ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 |
0.2 |
GO:0019046 |
release from viral latency(GO:0019046) |
| 0.1 |
0.5 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.1 |
0.4 |
GO:0009051 |
pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 |
0.4 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.1 |
0.1 |
GO:0044240 |
multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 |
2.0 |
GO:0014075 |
response to amine(GO:0014075) |
| 0.1 |
0.6 |
GO:0051016 |
barbed-end actin filament capping(GO:0051016) |
| 0.1 |
4.0 |
GO:0043542 |
endothelial cell migration(GO:0043542) |
| 0.1 |
1.2 |
GO:0007405 |
neuroblast proliferation(GO:0007405) |
| 0.1 |
0.1 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.1 |
1.3 |
GO:0048008 |
platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.1 |
0.2 |
GO:0060744 |
thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 |
0.2 |
GO:0006434 |
seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 |
1.0 |
GO:0045749 |
obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.1 |
0.3 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.1 |
0.1 |
GO:0071478 |
cellular response to radiation(GO:0071478) cellular response to light stimulus(GO:0071482) |
| 0.1 |
0.1 |
GO:0015791 |
polyol transport(GO:0015791) glycerol transport(GO:0015793) |
| 0.1 |
0.3 |
GO:0006386 |
transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.1 |
0.4 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.1 |
1.4 |
GO:0042147 |
retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 |
0.3 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.1 |
0.5 |
GO:0045329 |
amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.1 |
1.0 |
GO:0045104 |
intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 |
0.4 |
GO:0044245 |
polysaccharide digestion(GO:0044245) |
| 0.0 |
0.6 |
GO:0006662 |
glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 |
0.1 |
GO:0007538 |
primary sex determination(GO:0007538) |
| 0.0 |
0.5 |
GO:0001516 |
prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 |
0.2 |
GO:0031274 |
pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 |
0.2 |
GO:0009822 |
alkaloid catabolic process(GO:0009822) |
| 0.0 |
0.1 |
GO:0060024 |
negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) musculoskeletal movement, spinal reflex action(GO:0050883) rhythmic synaptic transmission(GO:0060024) |
| 0.0 |
0.5 |
GO:1903963 |
arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 |
0.1 |
GO:0002378 |
immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 |
0.9 |
GO:1901224 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 |
0.4 |
GO:0001510 |
RNA methylation(GO:0001510) |
| 0.0 |
0.3 |
GO:0070779 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 |
0.0 |
GO:0010040 |
response to iron(II) ion(GO:0010040) |
| 0.0 |
3.7 |
GO:0034249 |
negative regulation of translation(GO:0017148) negative regulation of cellular amide metabolic process(GO:0034249) |
| 0.0 |
0.1 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 |
0.5 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 |
0.1 |
GO:0097300 |
retinal cell programmed cell death(GO:0046666) necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 |
0.1 |
GO:0010834 |
obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 |
0.3 |
GO:0007176 |
regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 |
0.1 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.0 |
0.1 |
GO:0014041 |
regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 |
0.9 |
GO:0018208 |
peptidyl-proline modification(GO:0018208) |
| 0.0 |
0.1 |
GO:0060291 |
long-term synaptic potentiation(GO:0060291) |
| 0.0 |
1.0 |
GO:0006783 |
heme biosynthetic process(GO:0006783) |
| 0.0 |
3.5 |
GO:0030198 |
extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 |
2.9 |
GO:0030521 |
androgen receptor signaling pathway(GO:0030521) |
| 0.0 |
0.2 |
GO:0046642 |
negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 |
1.8 |
GO:0043484 |
regulation of RNA splicing(GO:0043484) |
| 0.0 |
0.8 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.0 |
4.0 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 |
0.1 |
GO:0007181 |
transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 |
2.4 |
GO:0007229 |
integrin-mediated signaling pathway(GO:0007229) |
| 0.0 |
0.1 |
GO:0043276 |
anoikis(GO:0043276) |
| 0.0 |
0.3 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.0 |
0.1 |
GO:0033144 |
negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) |
| 0.0 |
1.3 |
GO:0048477 |
oogenesis(GO:0048477) |
| 0.0 |
0.1 |
GO:0061084 |
regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) negative regulation of protein folding(GO:1903333) |
| 0.0 |
1.6 |
GO:0000289 |
nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 |
0.2 |
GO:0010890 |
positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 |
0.8 |
GO:0035050 |
embryonic heart tube development(GO:0035050) |
| 0.0 |
0.2 |
GO:0006658 |
phosphatidylserine metabolic process(GO:0006658) |
| 0.0 |
0.1 |
GO:0003056 |
regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 |
1.5 |
GO:0007193 |
adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 |
0.1 |
GO:0030037 |
actin filament reorganization involved in cell cycle(GO:0030037) actin filament reorganization(GO:0090527) |
| 0.0 |
0.2 |
GO:0006422 |
aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 |
0.1 |
GO:0007500 |
mesodermal cell fate determination(GO:0007500) |
| 0.0 |
0.5 |
GO:0031145 |
anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 |
0.5 |
GO:0060444 |
branching involved in mammary gland duct morphogenesis(GO:0060444) |
| 0.0 |
0.5 |
GO:0007035 |
vacuolar acidification(GO:0007035) |
| 0.0 |
0.3 |
GO:0045060 |
negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 |
0.7 |
GO:0008038 |
neuron recognition(GO:0008038) |
| 0.0 |
0.4 |
GO:0045494 |
photoreceptor cell maintenance(GO:0045494) |
| 0.0 |
0.2 |
GO:0043951 |
negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 |
0.1 |
GO:0032776 |
maintenance of DNA methylation(GO:0010216) DNA methylation on cytosine(GO:0032776) negative regulation of histone H3-K9 methylation(GO:0051573) C-5 methylation of cytosine(GO:0090116) |
| 0.0 |
0.2 |
GO:0060528 |
secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 |
0.1 |
GO:0045056 |
transcytosis(GO:0045056) |
| 0.0 |
0.2 |
GO:2000108 |
positive regulation of lymphocyte apoptotic process(GO:0070230) positive regulation of T cell apoptotic process(GO:0070234) positive regulation of thymocyte apoptotic process(GO:0070245) positive regulation of leukocyte apoptotic process(GO:2000108) |
| 0.0 |
0.8 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 |
0.2 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 |
0.3 |
GO:0009629 |
response to gravity(GO:0009629) |
| 0.0 |
0.2 |
GO:0043001 |
Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 |
0.8 |
GO:0046324 |
regulation of glucose import(GO:0046324) |
| 0.0 |
0.4 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 |
0.6 |
GO:0006739 |
NADP metabolic process(GO:0006739) |
| 0.0 |
0.7 |
GO:0033574 |
response to testosterone(GO:0033574) |
| 0.0 |
0.4 |
GO:0060638 |
mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 |
2.0 |
GO:0016197 |
endosomal transport(GO:0016197) |
| 0.0 |
0.2 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 |
0.3 |
GO:0019369 |
arachidonic acid metabolic process(GO:0019369) |
| 0.0 |
0.1 |
GO:0060662 |
tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 |
0.1 |
GO:0050678 |
regulation of epithelial cell proliferation(GO:0050678) |
| 0.0 |
0.4 |
GO:0006184 |
obsolete GTP catabolic process(GO:0006184) |
| 0.0 |
0.1 |
GO:0050747 |
positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 |
0.7 |
GO:0006081 |
cellular aldehyde metabolic process(GO:0006081) |
| 0.0 |
0.4 |
GO:0051412 |
response to corticosterone(GO:0051412) |
| 0.0 |
0.7 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
2.6 |
GO:1904668 |
positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 |
0.1 |
GO:0003416 |
endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.0 |
0.2 |
GO:0001502 |
cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 |
1.0 |
GO:0006903 |
vesicle targeting(GO:0006903) |
| 0.0 |
0.3 |
GO:0051298 |
centrosome duplication(GO:0051298) |
| 0.0 |
0.1 |
GO:0001514 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 |
0.2 |
GO:0045008 |
depyrimidination(GO:0045008) |
| 0.0 |
0.1 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) cytoplasmic translation(GO:0002181) cytoplasmic translational initiation(GO:0002183) |
| 0.0 |
0.1 |
GO:0007506 |
gonadal mesoderm development(GO:0007506) |
| 0.0 |
0.2 |
GO:0006379 |
mRNA cleavage(GO:0006379) |
| 0.0 |
0.1 |
GO:0014887 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 |
0.2 |
GO:0006702 |
androgen biosynthetic process(GO:0006702) |
| 0.0 |
0.8 |
GO:0010976 |
positive regulation of neuron projection development(GO:0010976) |
| 0.0 |
0.1 |
GO:0051927 |
obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.0 |
0.1 |
GO:0032223 |
negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 |
0.3 |
GO:0019370 |
leukotriene biosynthetic process(GO:0019370) |
| 0.0 |
0.4 |
GO:0043101 |
purine-containing compound salvage(GO:0043101) |
| 0.0 |
0.3 |
GO:0006493 |
protein O-linked glycosylation(GO:0006493) |
| 0.0 |
0.3 |
GO:0006891 |
intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 |
0.1 |
GO:0045065 |
cytotoxic T cell differentiation(GO:0045065) |
| 0.0 |
1.0 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
0.2 |
GO:0019228 |
neuronal action potential(GO:0019228) |
| 0.0 |
0.5 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 |
0.2 |
GO:0006750 |
glutathione biosynthetic process(GO:0006750) |
| 0.0 |
0.1 |
GO:0001649 |
osteoblast differentiation(GO:0001649) |
| 0.0 |
0.1 |
GO:0021675 |
nerve development(GO:0021675) |
| 0.0 |
0.1 |
GO:0001885 |
endothelial cell development(GO:0001885) |
| 0.0 |
0.1 |
GO:0016056 |
rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 |
0.4 |
GO:0050680 |
negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 |
0.3 |
GO:0046928 |
regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 |
0.0 |
GO:0006650 |
glycerophospholipid metabolic process(GO:0006650) |
| 0.0 |
0.6 |
GO:0071377 |
cellular response to glucagon stimulus(GO:0071377) |
| 0.0 |
0.2 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) |
| 0.0 |
0.2 |
GO:0006349 |
regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 |
0.0 |
GO:0010288 |
response to lead ion(GO:0010288) |
| 0.0 |
0.1 |
GO:0045351 |
type I interferon biosynthetic process(GO:0045351) |
| 0.0 |
0.4 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.0 |
0.3 |
GO:0015804 |
neutral amino acid transport(GO:0015804) |
| 0.0 |
0.1 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.0 |
0.1 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.0 |
0.1 |
GO:0042635 |
regulation of hair cycle(GO:0042634) positive regulation of hair cycle(GO:0042635) regulation of hair follicle development(GO:0051797) positive regulation of hair follicle development(GO:0051798) |
| 0.0 |
0.5 |
GO:0042246 |
tissue regeneration(GO:0042246) |
| 0.0 |
0.1 |
GO:1900087 |
traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.0 |
0.3 |
GO:0046470 |
phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 |
1.0 |
GO:0009636 |
response to toxic substance(GO:0009636) |
| 0.0 |
0.0 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.0 |
0.3 |
GO:0051899 |
membrane depolarization(GO:0051899) |
| 0.0 |
0.0 |
GO:0001829 |
trophectodermal cell differentiation(GO:0001829) |
| 0.0 |
0.3 |
GO:0001755 |
neural crest cell migration(GO:0001755) |
| 0.0 |
0.1 |
GO:0001916 |
positive regulation of T cell mediated cytotoxicity(GO:0001916) positive regulation of T cell mediated immunity(GO:0002711) |
| 0.0 |
0.2 |
GO:0006688 |
glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 |
0.1 |
GO:0045070 |
positive regulation of viral genome replication(GO:0045070) |
| 0.0 |
0.1 |
GO:0035272 |
exocrine system development(GO:0035272) |
| 0.0 |
0.0 |
GO:0016188 |
synaptic vesicle maturation(GO:0016188) |
| 0.0 |
0.1 |
GO:0043587 |
tongue development(GO:0043586) tongue morphogenesis(GO:0043587) |
| 0.0 |
0.5 |
GO:0034329 |
cell junction assembly(GO:0034329) |
| 0.0 |
0.2 |
GO:0045445 |
myoblast differentiation(GO:0045445) |
| 0.0 |
1.1 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.2 |
GO:0007202 |
activation of phospholipase C activity(GO:0007202) regulation of phospholipase activity(GO:0010517) positive regulation of phospholipase activity(GO:0010518) positive regulation of phospholipase C activity(GO:0010863) positive regulation of lipase activity(GO:0060193) regulation of phospholipase C activity(GO:1900274) |
| 0.0 |
1.0 |
GO:0018279 |
peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 |
0.1 |
GO:0034260 |
negative regulation of GTPase activity(GO:0034260) |
| 0.0 |
0.2 |
GO:0060334 |
regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 |
1.8 |
GO:0006470 |
protein dephosphorylation(GO:0006470) |