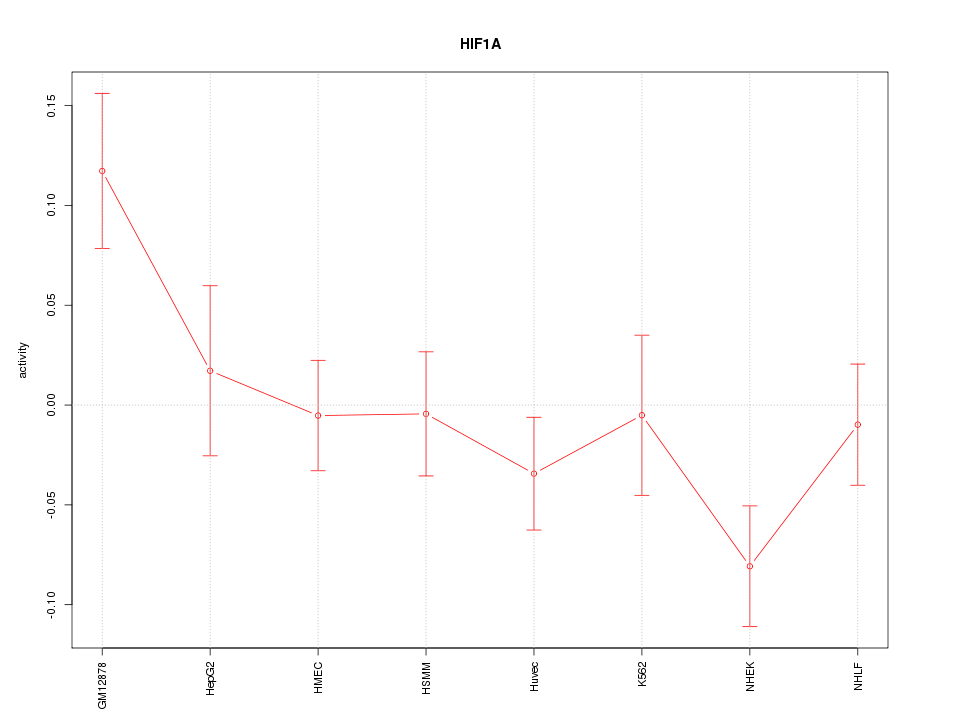
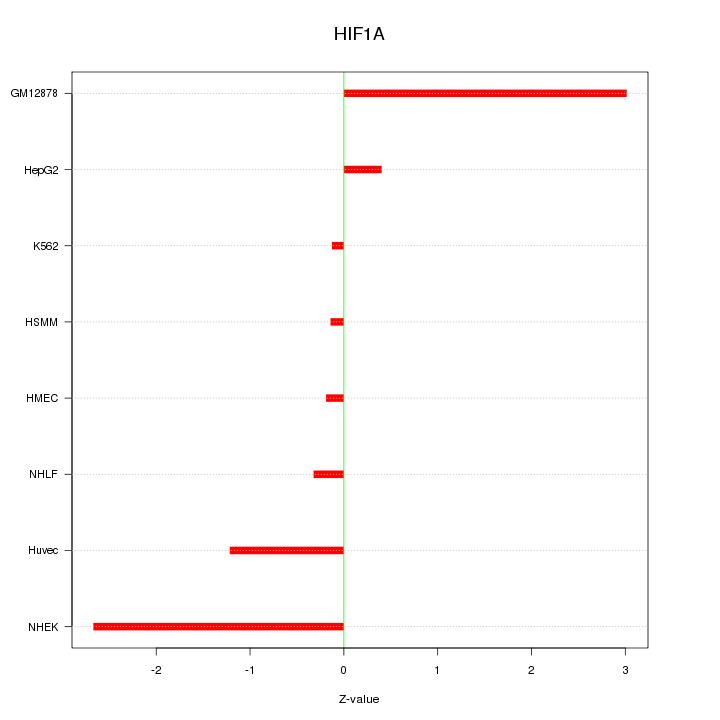
Motif ID: HIF1A
Z-value: 1.502
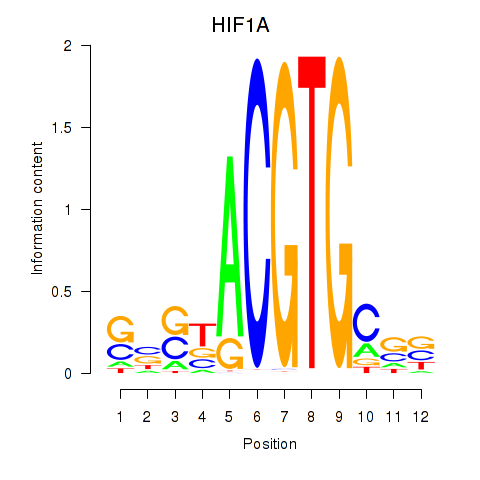
Transcription factors associated with HIF1A:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| HIF1A | ENSG00000100644.12 | HIF1A |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.9 | 3.5 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.8 | 2.3 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.6 | 1.8 | GO:1990170 | detoxification of cadmium ion(GO:0071585) stress response to cadmium ion(GO:1990170) |
| 0.6 | 1.7 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.5 | 2.6 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.5 | 1.4 | GO:0060318 | definitive erythrocyte differentiation(GO:0060318) |
| 0.4 | 1.5 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.3 | 1.3 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.2 | 1.9 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.2 | 1.4 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 1.0 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 | 0.7 | GO:0017014 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.2 | 0.3 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 | 0.5 | GO:0071679 | cell proliferation in midbrain(GO:0033278) commissural neuron axon guidance(GO:0071679) |
| 0.1 | 1.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.4 | GO:0007109 | obsolete cytokinesis, completion of separation(GO:0007109) protein hexamerization(GO:0034214) |
| 0.1 | 1.8 | GO:0010042 | response to manganese ion(GO:0010042) |
| 0.1 | 0.7 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 2.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 1.8 | GO:0006098 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.1 | 1.0 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 0.7 | GO:0045007 | depurination(GO:0045007) |
| 0.1 | 0.3 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.1 | 0.1 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 0.2 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.1 | 0.5 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 0.7 | GO:0006188 | IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.1 | 0.3 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.1 | 0.3 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.1 | 0.4 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 0.3 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 0.3 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.1 | 1.3 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.1 | 5.2 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.1 | 1.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 0.3 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 | 0.4 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.6 | GO:0015802 | basic amino acid transport(GO:0015802) |
| 0.0 | 0.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 1.0 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.7 | GO:0008634 | obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.0 | 0.3 | GO:0032367 | intracellular sterol transport(GO:0032366) intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.2 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.2 | GO:0043030 | regulation of macrophage activation(GO:0043030) |
| 0.0 | 0.2 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.1 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.7 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) embryonic cleavage(GO:0040016) |
| 0.0 | 0.2 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 | 0.1 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.2 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.4 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.3 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.0 | 2.0 | GO:0034339 | obsolete regulation of transcription from RNA polymerase II promoter by nuclear hormone receptor(GO:0034339) |
| 0.0 | 0.1 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 1.1 | GO:0046632 | alpha-beta T cell differentiation(GO:0046632) |
| 0.0 | 0.3 | GO:0006386 | transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 | 0.1 | GO:0016071 | mRNA metabolic process(GO:0016071) |
| 0.0 | 0.2 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.2 | GO:0033523 | histone monoubiquitination(GO:0010390) histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.1 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.4 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.7 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 0.3 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.3 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.0 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.3 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.0 | GO:0072106 | regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.0 | 0.1 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 1.6 | GO:0007611 | learning or memory(GO:0007611) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.6 | 2.2 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.4 | 1.4 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.3 | 0.8 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 1.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.2 | 1.9 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 2.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.2 | 2.1 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.2 | 1.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.7 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 4.6 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 2.1 | GO:0042645 | mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 3.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 1.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 5.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.4 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.2 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.7 | GO:0098796 | membrane protein complex(GO:0098796) |
| 0.0 | 0.1 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.0 | 0.2 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) exoribonuclease II activity(GO:0008859) |
| 0.8 | 4.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.8 | 2.3 | GO:0070361 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.7 | 2.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.6 | 1.8 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.6 | 1.7 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.5 | 2.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.4 | 1.2 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.4 | 1.6 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.4 | 1.2 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.3 | 1.4 | GO:0008907 | integrase activity(GO:0008907) |
| 0.2 | 1.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 0.7 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 1.9 | GO:0061659 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.2 | 0.7 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.2 | 1.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 1.4 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 1.8 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.7 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 1.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.7 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 1.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.5 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.1 | 0.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 1.0 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 0.2 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.1 | 0.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 2.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.6 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.2 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.2 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.5 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 1.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.2 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 0.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.4 | GO:0015605 | organophosphate ester transmembrane transporter activity(GO:0015605) |
| 0.0 | 0.8 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 2.1 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.1 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.3 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.6 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 0.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.3 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 2.6 | GO:0010843 | obsolete promoter binding(GO:0010843) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0046975 | RNA polymerase II core binding(GO:0000993) histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.5 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | SIG_IL4RECEPTOR_IN_B_LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.2 | SA_TRKA_RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.4 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.5 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.1 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |